- en Change Region
- Global Site
- Home
- Applications
- Life Sciences
- Live Cell Imaging
Live Cell Imaging
It is difficult to understate the importance of live cell microscopy – particularly fluorescence imaging – to modern cell biology research. Live cell fluorescence imaging allows for the dynamics and interactions of specific biomolecules to be observed over extended periods of time. Numerous technological innovations over the past few decades have contributed to the utility of live imaging, including the development of fluorescent proteins as genetically expressed and specific labels. Live cell imaging is typically performed using an inverted microscope to better accommodate medium-filled culture/imaging vessels.
Products for Live Cell Imaging
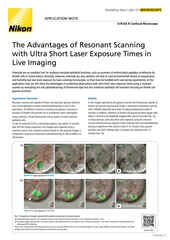
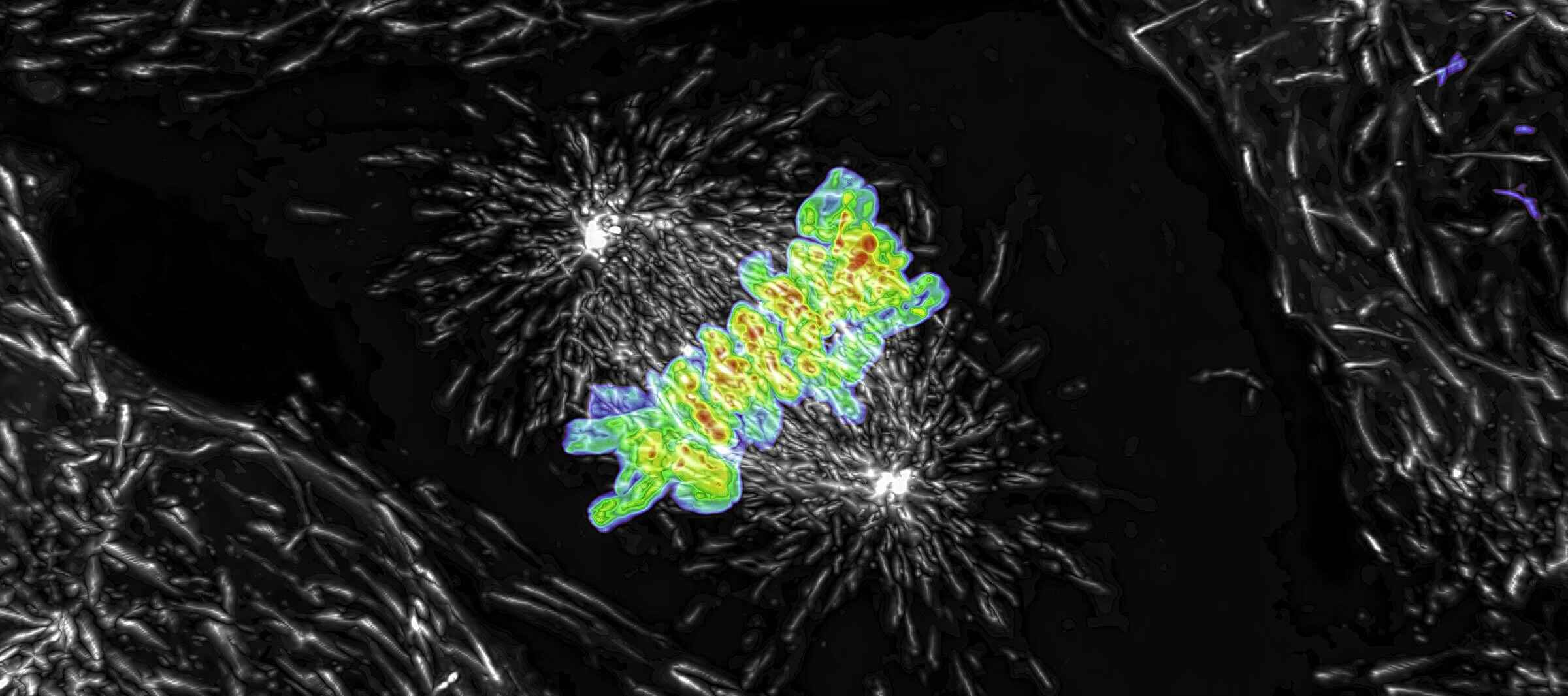
Maximum intensity projection of a Z stack collected of a live mitotic cell, acquired using the AX R resonant scanner operating at 15 FPS and a pixel resolution of 2048 x 1024. The objective lens was a CFI Plan Apo Lambda 60X Oil, 1.4 N.A.
The ECLIPSE Ti2-E motorized inverted microscope is the recommended microscope stand for live cell imaging – it can be combined with all of the confocal and super-resolution options detailed herein. The Perfect Focus System 4 (PFS4) is the industry’s leading hardware-based focus-locking technology, enabling long-term live cell imaging without focal drift and compatible with both glass and plastic culture vessels. An automatic water dispenser for water immersion objectives is also available.
The Ti2-LAPP modular illumination system allows a single microscope to be combined with up to five different illumination modules, with options available for TIRF, widefield epifluorescence, and photostimulation, including FRAP devices and digital micromirror devices (DMDs). It is possible to configure multiple TIRF illuminator modules on a single stand, allowing for simultaneous multi-channel imaging with the optimal TIRF angle for each laser line.
The BioPipeline LIVE High Content Imaging system is configured exclusively on the Ti2-E and includes a microscope incubation enclosure directly coupled to the cell culture incubator. It includes sample exchange robotics, which allows for automated high content imaging of up to 44 different vessels.
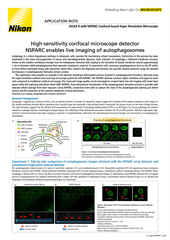
The AX / AX R confocal microscopes are Nikon’s point-scanning confocal solutions – with the AX R featuring a fast resonant-scanning system that supports video rate imaging of 30 frames per second (FPS) at 512 x 512-pixel resolution. Fast imaging is supported by the Denoise.ai software module, which automatically removes shot noise – the major source of noise in resonant scanning confocal images.
The ECLIPSE Ji digital inverted microscope utilizes the same Perfect Focus System (PFS) as the Ti2-E. By connecting a stage-top incubator, CO2 concentration, humidity, and temperature can be controlled to enable live cell imaging for extended periods of time. By integrating the AX / AX R with the ECLIPSE Ji, the above-mentioned high-speed acquisition, shot noise elimination using Denoise.ai, and imaging of thick samples are possible.
Yokogawa CSU series spinning disk confocal systems are popular solutions for live cell imaging. The CSU-W1 features wider inter-pinhole spacing, which reduces signal crosstalk between adjacent pinholes and enables deeper imaging in scattering samples such as organoids.
Super-resolution microscopy continues to become more refined and practical for live cell imaging applications. The Yokogawa CSU-W1 SoRa is a spinning disk confocal instrument that integrates an emission microlens disk to realize super-resolution via optical pixel reassignment.
●: included, ⚬: option
| ECLIPSE Ti2-E Inverted Microscope (Widefield Imaging)* |
AX R Resonant Confocal System |
CSU-X1 Spinning Disk Confocal Scanner |
CSU-W1 Spinning Disk Confocal Scanner |
Ti2-LAPP E-TIRF Illuminator |
CSU-W1 SoRa Spinning Disk Super-Resolution System |
|
|---|---|---|---|---|---|---|
| Relative Imaging Depth Limit | ~ 5 μm ~ 15 – 25 μm (with deconvolution) |
~ 100 – 500 μm | ~ 50 μm | ~ 50 – 100 μm | ~ 100 – 300 nm | ~ 50 – 100 μm |
| Supports Video Rate Imaging | Yes |
Yes (30 FPS with 512 x 512 scan) | Yes |
Yes (limited by camera system and disk rotation speed) |
Yes (limited by camera system) |
Yes (limited by camera system and disk rotation speed) |
| Field of View | 25 mm diagonal (circular) | 25 mm diagonal (square) | 10 x 7 mm (rectangular) | 17 x 16 mm (rectangular) | ~ 10 mm diagonal (circular) | 17 x 16 mm (rectangular) |
| Supported Imaging Modalities | ECLIPSE Ti2-E | AX R | CSU-X1 | CSU-W1 | Ti2-LAPP E-TIRF | CSU-W1 SoRa |
| Brightfield | yes | no | no | no | no | no |
| Confocal - Point-Scanning | no | yes | no | no | no | no |
| Confocal - Spinning Disk | no | no | yes | yes | no | yes |
| Darkfield | yes | no | no | no | no | no |
| Differential Interference Contrast (DIC) | yes | no | no | no | no | no |
| Nikon Advanced Modulation Contrast (NAMC) | yes | no | no | no | no | no |
| Phase Contrast | yes | no | no | no | no | no |
| Super-Resolution - Structured Illumination | no | no | no | no | no | no |
| Super-Resolution - Optical Pixel Reassignment Microscopy | no | no | no | no | no | yes |
| Total Internal Reflection Fluorescence (TIRF) | no | no | no | no | yes | no |
| Volume Contrast | yes | no | no | no | no | no |
| Widefield Fluorescence | yes | no | no | no | no | no |
| Compatible Microscope Stands | ECLIPSE Ti2-E | AX R | CSU-X1 | CSU-W1 | Ti2-LAPP E-TIRF | CSU-W1 SoRa |
| ECLIPSE Ti2-E Inverted | no | yes | yes | yes | yes | yes |
| ECLIPSE Ti2-A Inverted | no | no | yes | yes | no | yes |
| ECLIPSE Ti2-U Inverted | no | no | yes | yes | no | yes |
| ECLIPSE Ji Digital Inverted | no | yes | no | no | no | no |
| ECLIPSE Ni-E Upright | no | yes | yes | yes | no | no |
| ECLIPSE Ni-U Upright | no | no | yes | yes | no | no |
| ECLIPSE FN1 Upright | no | yes | yes | yes | no | no |
*This row refers solely to the use of the ECLIPSE Ti2-E for widefield imaging. The Ti2-E is the recommended microscope stand for use together with the other systems listed in the table.
Related Literature
Discussion of Live Cell Imaging
The Art and Science of Live Cell Imaging
The Nikon Silicone Immersion Series objective lenses. Each of these lenses provides high NA, long working distance, and a correction collar with temperature-dependent settings.
Live cell imaging requires users to carefully balance the imaging conditions – the aim is to collect the minimal essential data, which minimizes perturbance of the model system. While it can be tempting to use more illumination power, longer camera exposures, etc. to obtain higher signal-to-noise ratio (SNR) images, it’s important to understand that phototoxicity can compromise the physiological integrity and continued viability of the system. Furthermore, photobleaching imposes a practical limit to how many photons can be extracted from each fluorophore over the course of the experiment.
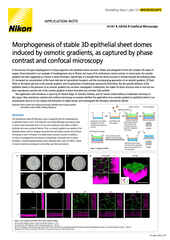
Another factor to consider is choice of objective lens. High numerical aperture (NA) oil objective lenses may work well for imaging features near the coverslip in flat cells, but suffer from spherical aberration the deeper one images into the sample. This is due to the lower refractive index (RI) of the cell/culture medium compared to glass/oil. For deeper and 3D imaging applications, it is recommended to use objectives that utilize immersion media more closely matching the RI of the imaging environment, such as water or silicone.
The CFI Apochromat Lambda S Objective Series includes a number of water immersion objective lenses, including the CFI Plan Apochromat IR 60XC WI, which has the highest NA for a water immersion objective at NA = 1.27 that we are aware of. Nikon has more recently introduced its Silicone Immersion Series objective lenses. Silicone has an RI of about 1.4, more closely matching the cellular environment while allowing for higher NAs than water immersion. These objectives are suited to imaging 3D culture systems such as organoids.
Minimizing Phototoxicity and Photobleaching in Live Cells

Hardware triggering significantly improves the overall experiment time. In most of these examples, the camera exposure time was longer than the camera readout time.
a The last row presents a comparison for which the camera exposure time was the same as its readout time, and the camera was able to run in overlapped (100% duty cycle) mode.
The intense irradiation required by fluorescence-based imaging techniques is inherently harmful to cell health, especially with higher-energy (blue-shifted) wavelengths. For this reason, the use of red-to-near-infrared fluorophores are recommended, though the continued use of popular green fluorophores such as EGFP is not overly-prohibitive and continues to work well for many multicolor imaging applications.
Most microscopes utilize software triggering of system devices. However, this can be somewhat slow due to the relative inaccuracy of the computer clock and the required checks and callbacks of device states. Conversely, hardware triggering uses the high accuracy camera pixel clock to coordinate devices while bypassing device callbacks, thus maximizing system speed while minimizing photon dose. Triggered devices may include Z piezo stages, laser/LED illuminators, and even custom devices featuring I/O communication capability.
Triggering also allows for more complex illumination patterns. For example, it is possible using the Nikon NIS-Elements software to illuminate the sample only when all pixels of a sCMOS camera are exposing (accounting for the effect of the rolling shutter) – decreasing photon dose and photobleaching compared to continuous illumination and free-running image acquisition. It is also possible to pulse illumination on the micro-second time scale during the exposure with various LED and laser illuminators. Pulsed illumination on this time scale allows fluorophores excited to a triplet state extra time to relax to the ground state, and thus help avoid major photobleaching pathways.
Using Deep Learning-based Artificial Intelligence to Improve Live Cell Imaging
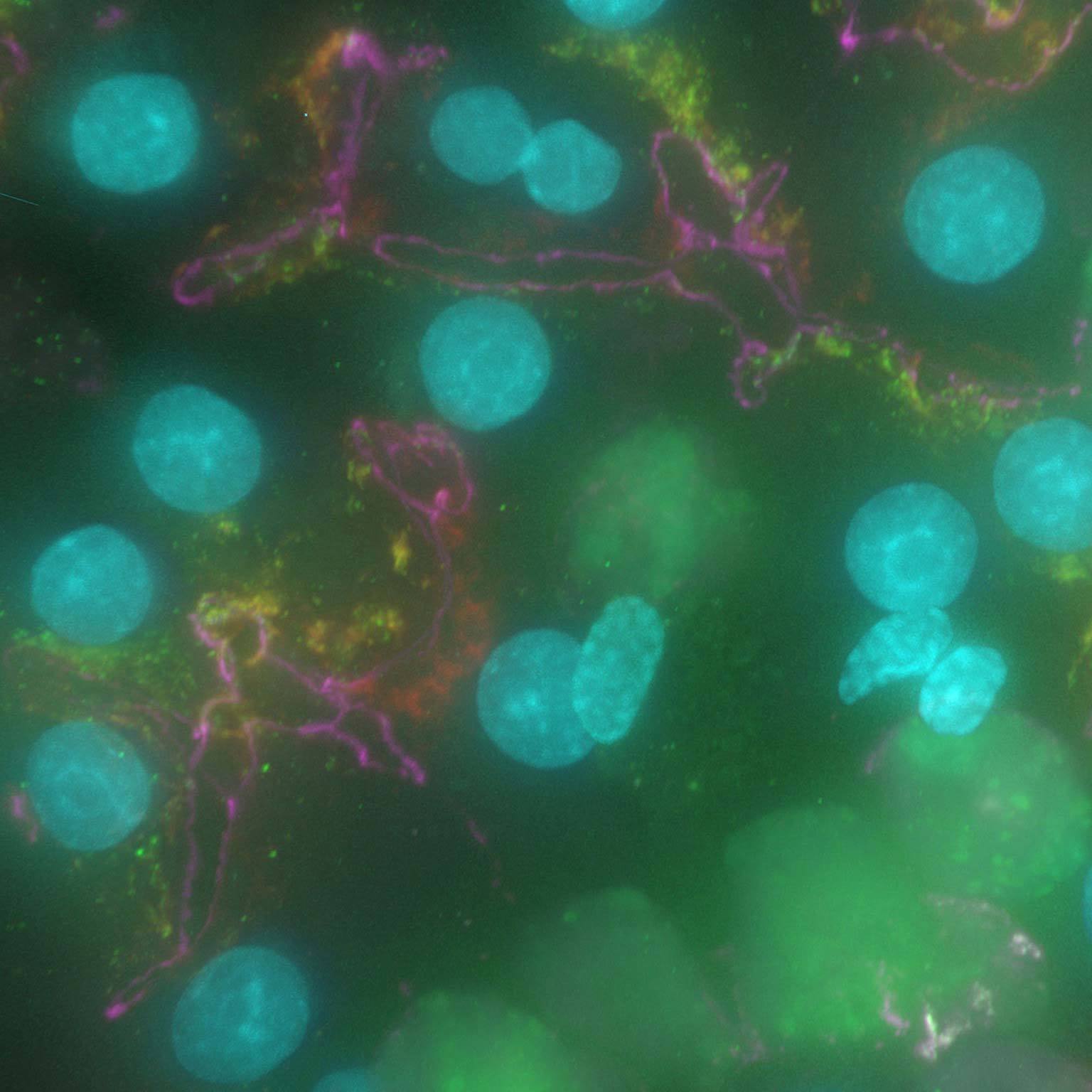
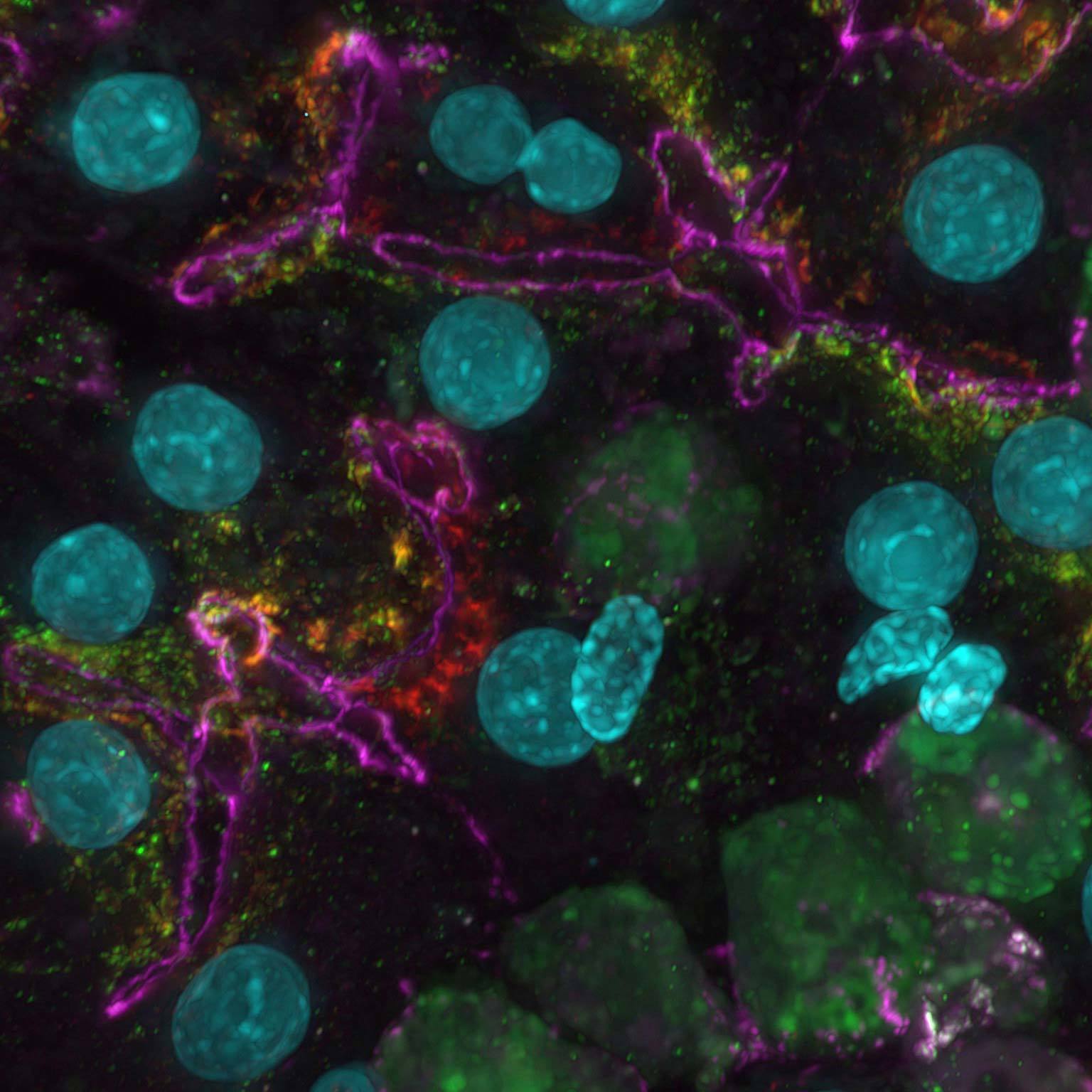
Widefield data can be contaminated by scattered and out-of-focus light, but AI-based tools can recover high contrast images by removing noise and blur.
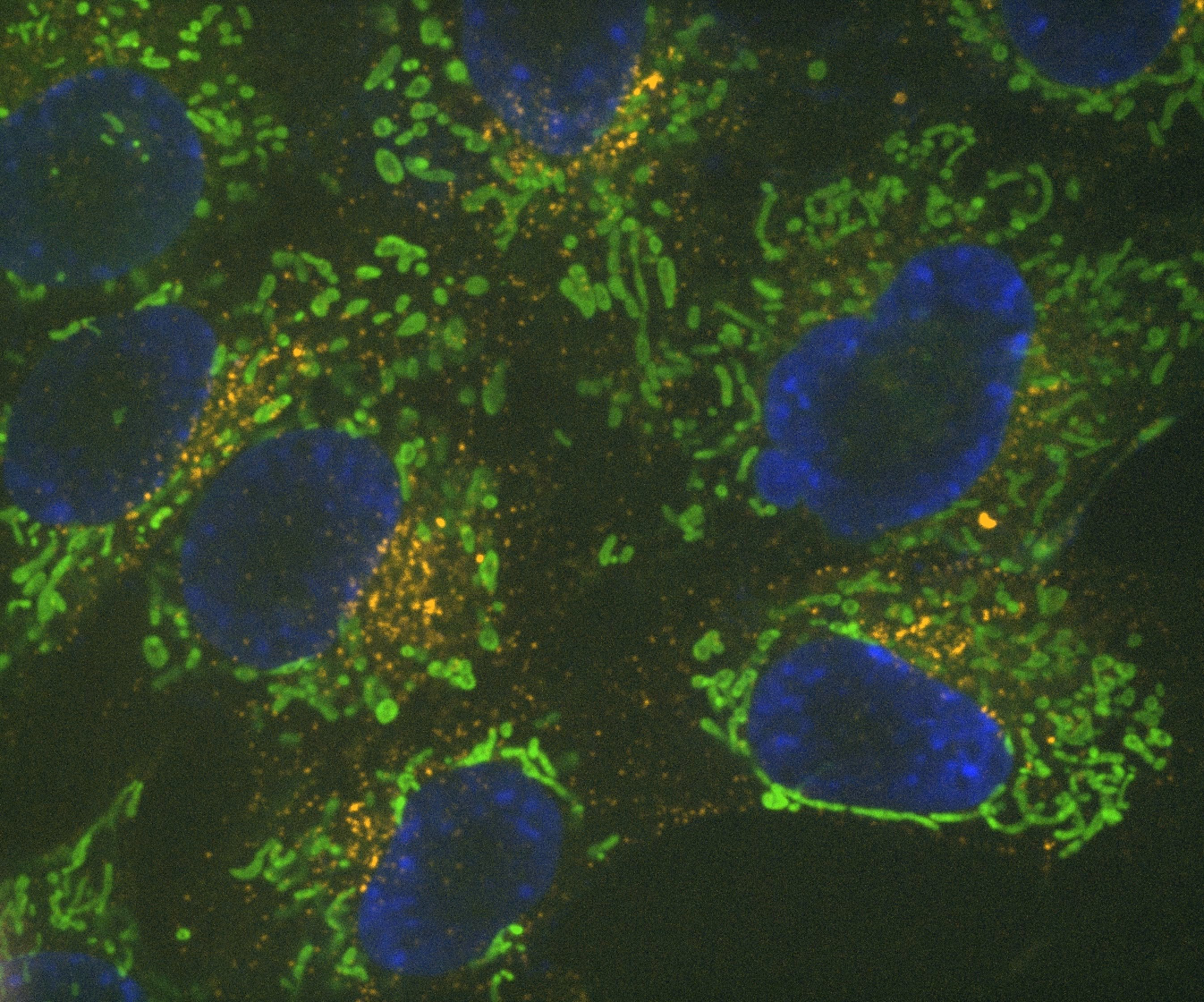
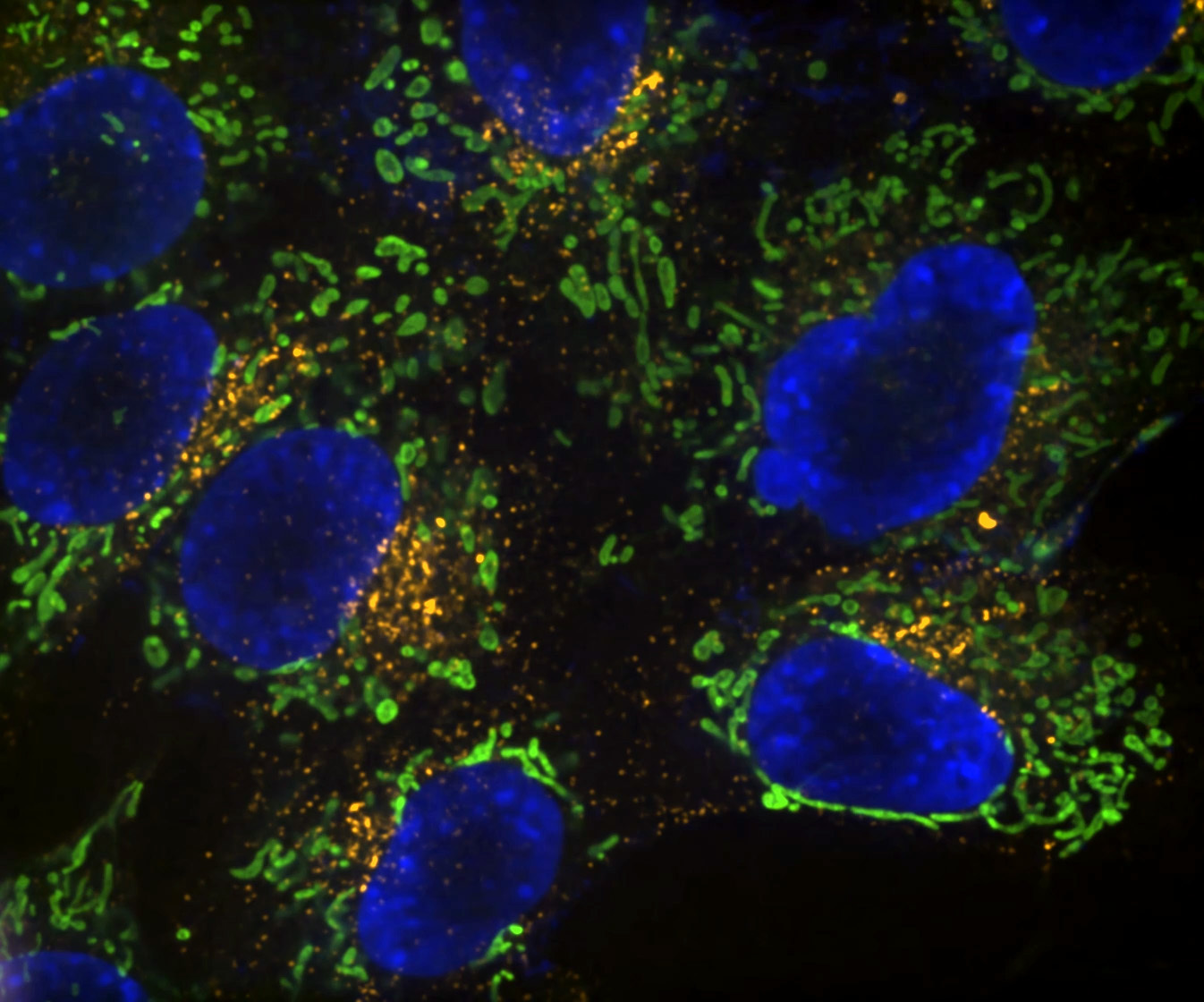
Denoise.ai can be applied to remove the shot noise component of images while leaving the underlying structure and intensity values undisturbed.
The advent of artificial intelligence (AI) methods for microscopy has opened up new possibilities for live cell imaging with reduced photon dose. Methods based on deep learning are proving suited towards assays based on images of biological specimens. Nikon is committed to the development of stable software solutions based on deep learning for various applications – available as NIS.ai modules for our NIS-Elements software.
Clarify.ai provides automatic removal of out-of-focus blur from standard widefield fluorescence images. This module is pre-trained at our factory and does not require manual user-based adjustment of the final image, removing the opportunity for subjectivity. Clarify.ai thus allows for optical sectioning to be performed on single plane widefield images acquired at the maximum system rate, not requiring data from multiple planes as with 3D iterative deconvolution of widefield data.
Denoise.ai is another pre-trained module and used to remove Poisson noise (shot noise) from confocal images in real time. This allows for reduced exposure times and is advantageous when used in conjunction with resonant scanning images acquired using the AX R confocal microscope, where shot noise is the predominant source of noise.
Nikon also offers the Convert.ai module for predicting image features that are typically detected in a fluorescence channel, with the predicted signal based on a corresponding brightfield image (e.g., prediction of DAPI nuclear staining patterns from DIC image data). This allows for elimination of fluorescence imaging channels, avoiding the intense illumination demanded by fluorescence techniques. Brightfield imaging techniques are inherently less phototoxic.
Enhance.ai can be trained to predict details in low signal-to-noise (SNR) images, effectively improving SNR and allowing for imaging with reduced photon dose and photobleaching.
Which Contrasting Technique is Right for Your Research?
Widefield fluorescence image of a mouse kidney glomerulus in a 16 mm thick tissue section before (a) and after (b) 3D iterative deconvolution in the NIS-Elements software.
For relatively flat two-dimensional samples, such as adherent cells cultured in vitro, widefield fluorescence imaging may be sufficient. Compatible use-cases and sample types are much broader when combining widefield imaging with deconvolution analysis or automatic deblurring using the Clarify.ai software module.
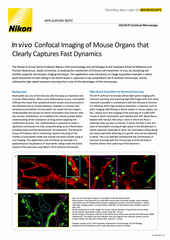
Confocal imaging becomes necessary when imaging samples that are more than ~20 μm thick but are also useful for high-resolution imaging of samples even just a few μm thick. Spinning disk confocal instruments are fast and suited to samples up to ~50 μm (with the CSU-W1 able to image deeper than other models due to greater inter-pinhole spacing). Both systems are applicable towards imaging different types of 3D cell culture systems, such as organoids and spheroids.
Point-scanning confocal imaging is able to image deeper, up to several hundred μm. The resonant scanner featured on the AX R confocal is able to provide video rate imaging and is intended for combination with Denoise.ai for real-time shot noise removal. Unlike spinning disk systems, the AX R features a continuously adjustable pinhole for optimization of optical sectioning and resolution with any compatible objective lens.
Total internal reflection fluorescence (TIRF) imaging is an optical sectioning technique that allows for exclusive observation of cell features occurring within a few hundred nm of the cell-coverglass interface. This provides a improvement in SNR, allowing for the use of lower laser power to protect cell health and/or maximize imaging speed. Nikon’s CFI Apochromat TIRF Series objective lenses also feature a very high NA of 1.49.
Super-resolution microscopy techniques are for when optical resolution beyond the diffraction limit is specifically required to resolve the necessary details. Live cell super-resolution microscopy has generally been challenging due to the tradeoffs demanded of these varied techniques to improve resolution. The Yokogawa CSU-W1 SoRa is a super-resolution system implemented in the context of a spinning disk confocal system, allowing for imaging at speeds matching the CSU-W1 alone (assuming sufficient signal).
While fluorescence imaging techniques are powerful, allowing for multiplexed detection of distinct molecular targets, brightfield imaging techniques such as phase contrast and differential interference contrast (DIC) provide detailed cell images with significantly lower photon dose. While such techniques may lack the required molecular-specificity, it is good practice to acquire occasional brightfield images in order to help assess cell health over the course of the experiment.
Glossary
- Compatible Microscope Stands
- This refers to the Nikon microscope stand models that are compatible with each system.
- Field of View
- The field of view of the system, also referred to as the field number, is the diameter of the imaging area at a nominal 1X magnification.
- Relative Imaging Depth Limit
- This indicates the approximate Z (axial) depth range within which the indicated system can deliver images with sufficient optical sectioning quality and signal-to-noise. This value can be quite variable and depends heavily on the optical properties of the specimen and vessel, as well as the labeling.
- Supported Imaging Modalities
- This refers to the various microscopy imaging techniques provided by each system. Note that almost all the imaging modalities supported by the ECLIPSE Ti2-E inverted microscope can still be accessed when it is used as a base for any of the other systems listed in this table.
- Supports Video Rate Imaging
- “Video rate” is traditionally defined as about 30 frames per second (FPS). Optimal imaging rate depends on exact application, and may be faster or slower than 30 FPS. EM-CCD cameras can typically image up to 60 FPS (full frame) and sCMOS cameras up to 40-100 FPS (full frame).
- Home
- Applications
- Life Sciences
- Live Cell Imaging