- en Change Region
- Global Site
Application Notes
Automatic isolation of pollen using NIS-Elements General Analysis (GA) and JOBS imaging workflow tools
November 2023
Single-cell analysis is important for understanding how individual cells function and how they respond as a population. However, depending on the sample, it may not be possible to isolate specific cells while they are alive due to incompatibility with the sheathe fluid or flow rate of a cell-sorting device. Additionally, during single-cell isolation via cell sorter or fine capillary, the position and environment of cells change before and after sorting, making it difficult to observe and evaluate cells over time. While there are isolation methods based on antibiotics, these cannot be used for some of cell types such as pollen.
To bypass these problems, Dr. Yoko Mizuta et al., of the Institute of Transformative Bio-Molecules, Nagoya University, use the General Analysis (GA) and JOBS of NIS-Elements imaging software to efficiently isolate target cells by automated, infrared laser-mediated disruption of pollen grains within pollen populations. JOBS combines the individual functions of NIS-Elements, such as microscope control, image acquisition, image processing, and external device control, allowing users to create their own imaging workflows, and GA enables the automation of image analysis. This application note introduces automatic pollen isolation using GA and JOBS, based on the research of Dr. Mizuta et al.
Keywords: pollen, single-cell analysis, automated isolation, JOBS, GA
Overview of automatic pollen isolation
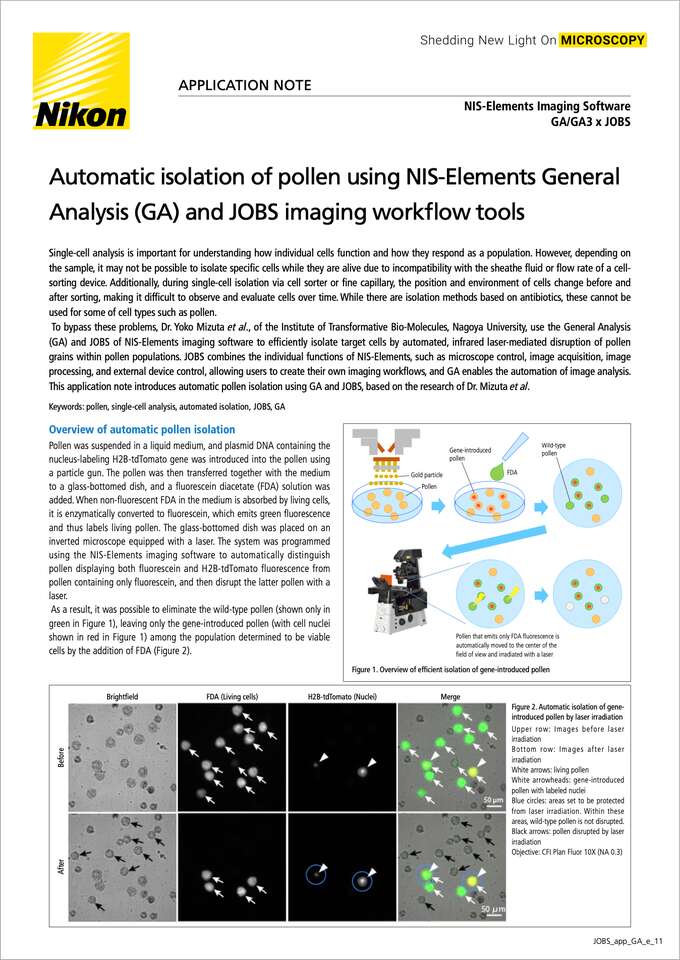
Pollen was suspended in a liquid medium, and plasmid DNA containing the nucleus-labeling H2B-tdTomato gene was introduced into the pollen using a particle gun. The pollen was then transferred together with the medium to a glass-bottomed dish, and a fluorescein diacetate (FDA) solution was added. When non-fluorescent FDA in the medium is absorbed by living cells, it is enzymatically converted to fluorescein, which emits green fluorescence and thus labels living pollen. The glass-bottomed dish was placed on an inverted microscope equipped with a laser. The system was programmed using the NIS-Elements imaging software to automatically distinguish pollen displaying both fluorescein and H2B-tdTomato fluorescence from pollen containing only fluorescein, and then disrupt the latter pollen with a laser.
As a result, it was possible to eliminate the wild-type pollen (shown only in green in Figure 1), leaving only the gene-introduced pollen (with cell nuclei shown in red in Figure 1) among the population determined to be viable cells by the addition of FDA (Figure 2).
(1) 画像取得範囲(35mmディッシュの内接)とレーザー照射時間を設定。
References
Target pollen isolation using automated infrared laser-mediated cell disruption
Ikuma Kaneshiro, Masako Igarashi, Tetsuya Higashiyama and Yoko Mizuta
Quantitative Plant Biology , Volume 3 , 2022 , e30
