- en Change Region
- Global Site
- Home
- Imaging Applications
- Life Sciences
- Nano-Scale Imaging
Nano-Scale Imaging
For hundreds of years the resolution of light microscopy was fundamentally limited by diffraction, with ~200 nanometers (nm) being regarded as the approximate limit in XY. This may be sufficient for observing single cells and many sub-cellular features, but it is insufficient for resolving fine sub-organelle scale details and single biomolecules, which often have dimensions in the single nm.
Recent history has witnessed the invention of numerous “super-resolution” optical imaging methods, exploiting different strategies for circumventing the limit imposed by diffraction. The importance of super-resolution was recognized with the 2014 Nobel Prize in Chemistry and has already contributed to several discoveries, such as the structure of the axonal periodic scaffold.
Products for Nano-Scale Imaging
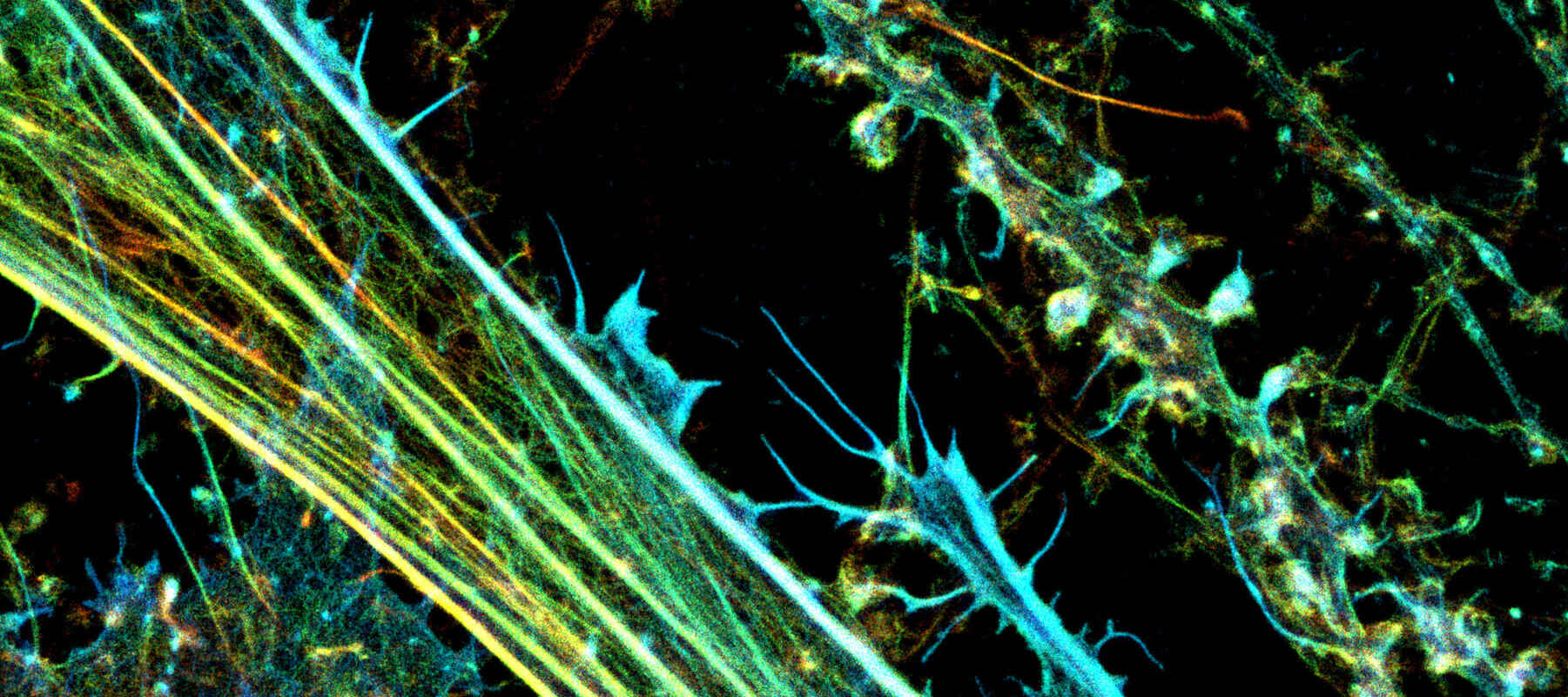
3D-STORM image of a neuronal culture with fluorescently labeled actin (color-coded for depth), acquired using a Nikon N-STORM system by Prof. Christophe Leterrier (CR1 CNRS).
The Nikon N-STORM Super-Resolution Microscope System provides the highest resolution of the systems offered by Nikon, approximately an order of magnitude greater compared to widefield epifluorescence in XYZ. The N-STORM is configured on the Ti2-E inverted microscope, which provides high stability thanks to the Perfect Focus System 4 (PFS4) focus-locking system, which is also utilized for fine axial translation for our Z-stacking STORM imaging mode, allowing for volumes up to ~5 µm thick to be imaged. Conventional total internal reflection fluorescence (TIRF) imaging is also possible using the N-STORM.
The N-SIM S Super-Resolution Microscope System uses a spatial light modulator to realize fast-switching of interference patterns, providing a speed boost for both 9-image (2D-SIM, TIRF-SIM) and 15-image (3D-SIM) modes. 9-image SIM can be performed at rates of up to 15 frames per second (FPS) – compatible with many live cell imaging applications. TIRF-SIM provides an additional resolution increase in (XY), along with the optical sectioning of TIRF microscopy. The N-SIM E Super-Resolution Microscope System is a more cost-effective SIM model for 3D-SIM imaging, and provides the same resolution improvement as the N-SIM S.
The Yokogawa CSU-W1 SoRa Super-Resolution Spinning Disk Confocal System is a spinning disk confocal instrument that integrates an emission microlens disk to realize super-resolution by optical pixel reassignment. Since the CSU-W1 SoRa uses a spinning disk design, image acquisition is fast.
The Abberior STEDYCON Super-Resolution Microscope System provides confocal and STED imaging together in a compact plug-and-play scan head that’s alignment-free and easily mounted on a standard microscope C-mount. The STEDYCON includes its own built-in focus-locking system and high efficiency avalanche photo detectors, with quantum efficiency up to 69% (at 650 nm).
●: included, ⚬: option
| N-STORM Super-Resolution Microscope System |
N-SIM S Super-Resolution Microscope System |
N-SIM E Super-Resolution Microscope System |
Yokogawa CSU-W1 SoRa Super-Resolution Spinning Disk Confocal System |
Abberior STEDYCON Super-Resolution Microscope System |
|
|---|---|---|---|---|---|
| Relative Optical Resolution Limit | ~20 nm (XY)* ~50 nm (Z)* |
~115 nm (XY)* ~86 nm (XY; TIRF-SIM mode)* ~269 nm (Z; 3D-SIM mode)* |
~115 nm (XY)* ~269 nm (Z; 3D-SIM mode)* |
~120 nm (XY)* | ~30 - 40 nm (XY)* |
| Relative Imaging Depth Limit | ~ 5 μm | ~ 10 – 20 μm | ~ 10 – 20 μm | ~ 50 – 100 μm | ~ 100 μm |
| Supported Image Acquisition Rate | ~0.1 FPS (up to 500 Hz acquisition of single image frames) |
~15 FPS (2D-SIM and TIRF-SIM only) |
~1 FPS (all modes) |
~30+ FPS (limited only by signal-to-noise, disk rotation speed, and camera readout rate) |
1.1 FPS (at 512 x 512-pixel resolution) |
| Supported Spectral Channels | 3 | 6 | 3 | 6 | 2 |
| Maximum Image Acquisition Area | 80 x 80 μm (using 100X objective) |
66 x 66 μm (using 100X objective) |
66 x 66 μm (using 100X objective) |
61 x 57 μm (using 100X objective with 2.8X magnification lens) |
90 x 80 μm (using 100X objective) |
| Compatible Microscope Stands | N-STORM | N-SIM S | N-SIM E | CSU-W1 SoRa | STEDYCON |
| ECLIPSE Ti2-E Inverted | yes | yes | yes | yes | yes |
| ECLIPSE Ti2-A Inverted | no | no | no | yes | no |
| ECLIPSE Ti2-U Inverted | no | no | no | yes | no |
| ECLIPSE Ni-E Upright | no | no | no | no | yes |
| FN1 Upright | no | no | no | no | yes |
*These values are provided as an approximate reference. Resolution performance will vary depending on exact conditions. Resolution estimates are not provided for dimensions where improvement is not expected.
Related Literature
Discussion of Nano-Scale Imaging
Finding the Super-Resolution Method That’s Right for You
Quantitative spine analysis with Z-stack of 3D-SIM, 35 steps, Z range: 4.2 um
Exposure time 100 ms, 120 sec intervals
Time-lapse of 11 times
Excitation wavelength 488 nm
Sample information: Dendritic spine in mouse hippocampal neuron expressing GFP
Movie courtesy of: Drs. Yutaro Kashiwagi and Shigeo Okabe, Department of Cellular Neurobiology, Graduate School of Medicine and Faculty of Medicine, The University of Tokyo.
Super-resolution imaging techniques provide resolution improvement past the diffraction limit, but each technique also has its own limitations and trade-offs that should be well understood in order to determine which is best suited towards your research.
First and foremost, super-resolution acquisition rates are generally slower than traditional techniques. Imaging with N-STORM is difficult for application in live cells due to the slower acquisition rate (almost universally requiring more than one second per frame). STORM and other SMLM techniques provide the greatest resolution improvement, but they are compatible with a limited number of fluorophores, many of which require specialized buffer systems that aren’t compatible with live cells.
The N-SIM S and Yokogawa CSU-W1-SoRa are better suited for super-resolution live cell imaging as they support faster imaging rates (15 FPS for N-SIM S, higher for CSU-W1-SoRa) and can be used with conventional fluorophores without specific buffer systems.
STED imaging with the STEDYCON system is the best choice among the options supported by Nikon for super-resolution imaging in thicker samples, such as tissue sections. This is due to the use of point-scanning acquisition with longer wavelengths. While the choice of fluorophore is more restrictive, similar to the N-STORM, there is no requirement for a specialized imaging buffer system.
High-Performance Objective Lenses for Super-Resolution
Nikon automated correction collar objective lens mounted on a Ti2-E inverted microscope.
One of the most critical factors with regard to maximizing optical resolution is the choice of objective lens. This applies equally to super-resolution as to other imaging techniques.
The numerical aperture (NA) of the objective lens should be as high as possible while still using an appropriate immersion medium. The Nikon Super-Resolution Series Microscope Objective Lenses includes the CFI SR HP Apochromat TIRF 100XC Oil (oil immersion, NA = 1.49), CFI SR HP Plan Apochromat Lambda S 100XC Sil (silicone immersion, NA = 1.35), and the CFI SR Plan Apochromat IR 60XC WI (water immersion, NA = 1.27). These objectives provide some of the highest-level NAs in their respective classes.
Select Super-Resolution Series objectives are available with an automatic correction collar, allowing for very fine-tuning of spherical aberration correction without manual adjustment, ensuring the highest-level resolution and 3D imaging performance possible.
Enhanced Resolution – Pushing the Limits of Traditional Technologies
Point-scanning confocal images of cochlear cilia. XY and XZ views with (right) and without (left) enhanced resolution settings applied.
Sometimes the resolution needed to answer your question is not too far beyond the conventional, and thus may not necessitate some of the trade-offs demanded by super-resolution methods. In such an instance we recommend that you consider the potential of what we refer to as “enhanced resolution” confocal imaging.
What is enhanced resolution confocal imaging? The key point to understand is that the traditional confocal microscope is already capable of a certain degree of resolution improvement beyond what’s possible with a typical widefield microscope, with a theoretical limit of ~140 nm (XY) being well described. However, reaching this limit requires the use of an infinitely small pinhole aperture. Fortunately, in practice a pinhole size of ~0.5 Airy Units can be combined with post-acquisition 3D iterative deconvolution to approach this limit, with ~160 nm resolution (XY) being a realistic target for a range of imaging conditions.
The Nikon AX / AX R point-scanning confocal microscopes are capable of enhanced resolution imaging. Advantages include being able to precisely tune detection for most visible spectrum fluorophores and supporting simultaneous acquisition of up to four spectral channels. The pinhole is continuously variable and has a hexagonal (rather than square) shape, allowing for more precise adjustment. With the AX R resonant scanning confocal model, it’s possible to image at video rate (30 FPS) and faster while still benefitting from enhanced resolution.
Glossary
- Compatible Microscope Stands
- This refers to the Nikon microscope stand models that are compatible with each system.
- Maximum Image Acquisition Area
- The largest field of view (measured in the sample space) supported by the technique using the specified objective lens magnification factor.
- Relative Imaging Depth Limit
- This indicates the approximate Z (axial) depth range within which the indicated system can deliver images with sufficient optical sectioning quality and signal-to-noise. This value can be quite variable and depends heavily on the optical properties of the specimen and vessel, as well as the labeling.
- Relative Optical Resolution Limit
- The practical resolution limit of the given system. The value for lateral (XY) resolution is typically different than that for axial (Z) resolution. If a Z-resolution value is not given, then no improvement is provided in that dimension.
- Supported Image Acquisition Rate
- Rates are shown as frames per second (FPS).
- Supported Spectral Channels
- This refers to the number of possible spectral (color) channels that are supported (assuming one laser line for each type of fluorophore).
- Home
- Imaging Applications
- Life Sciences
- Nano-Scale Imaging