- en Change Region
- Global Site
- Home
- Applications
- Life Sciences
- Deep Learning in Microscopy
Deep Learning in Microscopy
Artificial intelligence (AI) methods are gaining in popularity for microscopy data analysis. Deep learning (DL) is a type of machine learning (ML) that uses an artificial neural network (ANN) for learning. ANNs are so named because of the similarities with our own brain – each “neuron” (node) in the network receives inputs from multiple other nodes. The training process changes the weight of these connections to best predict the results expected from a given input.
Products for Deep Learning in Microscopy
DL approaches can be advantageously applied to a variety of image analysis tasks. Nikon’s NIS.ai software modules for the NIS-Elements software leverage DL for various image analyses. NIS.ai modules can be combined with other software tools as part of automated imaging and analysis pipelines using our JOBS experiment wizard and General Analysis 3 (GA3) module, respectively.
●: included, ⚬: option
| Autosignal.ai | Clarify.ai | Convert.ai | Denoise.ai | Enhance.ai | Segment.ai | |
|---|---|---|---|---|---|---|
| Function | Automatically determines optimal laser power and gain for point-scanning confocal imaging. | Automatically removes out-of-focus fluorescence signal from widefield fluorescence images. | Predicts images in a given channel from patterns identified in a transmitted light channel. | Automatically removes noise from the image. | Predicts an image with improved signal-to-noise from a low signal image. | Segments complex image features based on patterns identified from user-guided pre-training. |
| Pre-training Required | no | no | yes | no | yes | yes |
| Compatible Nikon Systems | AX / AX R Confocal Microscopes | All | All | AX R Confocal Microscopes and CMOS camera-based systems | All | All |
| Software Packages | Autosignal.ai | Clarify.ai | Convert.ai | Denoise.ai | Enhance.ai | Segment.ai |
| NIS-Elements C | Optional | Optional | Optional | Optional | Optional | Optional |
| NIS-Elements AR | no | Optional | Optional | Optional | Optional | Optional |
| NIS-Elements AR ML | no | Optional | Optional | Optional | Optional | Optional |
| NIS-Elements AR Passive | no | Optional | Optional | Optional | Optional | Optional |
| NIS.ai Module | yes | Optional | Optional | yes | Optional | Optional |
| Bundled with deconvolution modules | no | Yes | no | no | no | no |
Related Literature
Discussion of Deep Learning in Microscopy
Understanding the Benefits and Limitations of Deep Learning for Microscopy
DIC, ground truth, and Convert.ai reconstruction images of a spheroid in culture. Convert.ai was able to adequately reconstruct the position and shape of the fluorescence label, if not every exact detail.
While deep learning-based algorithms and software hold much promise for supporting optical microscopy image analysis – it is important to understand that such approaches are not a “cure-all,” but rather different ways to approach a problem. Deep learning (DL) algorithms, which work by inferring correlations between data features (which may not necessarily be obvious or intuitive), will be more well-suited to some problems than classical approaches. However, often the greater benefit of artificial intelligence (AI) for microscopy is acceleration of image analysis.
Supervised DL algorithms, as used by Nikon’s NIS.ai AI modules, require extensive data for training the neural network. While Nikon pre-trains some AI modules, including Denoise.ai, Clarify.ai, and Autosignal.ai, others will need to be trained using the specific acquisition conditions for a given experiment. This process can be lengthy, but once completed it will allow for fast analysis of new data. It is important to understand that the results are not the ground truth. Ultimately, this limits the appropriate applications.
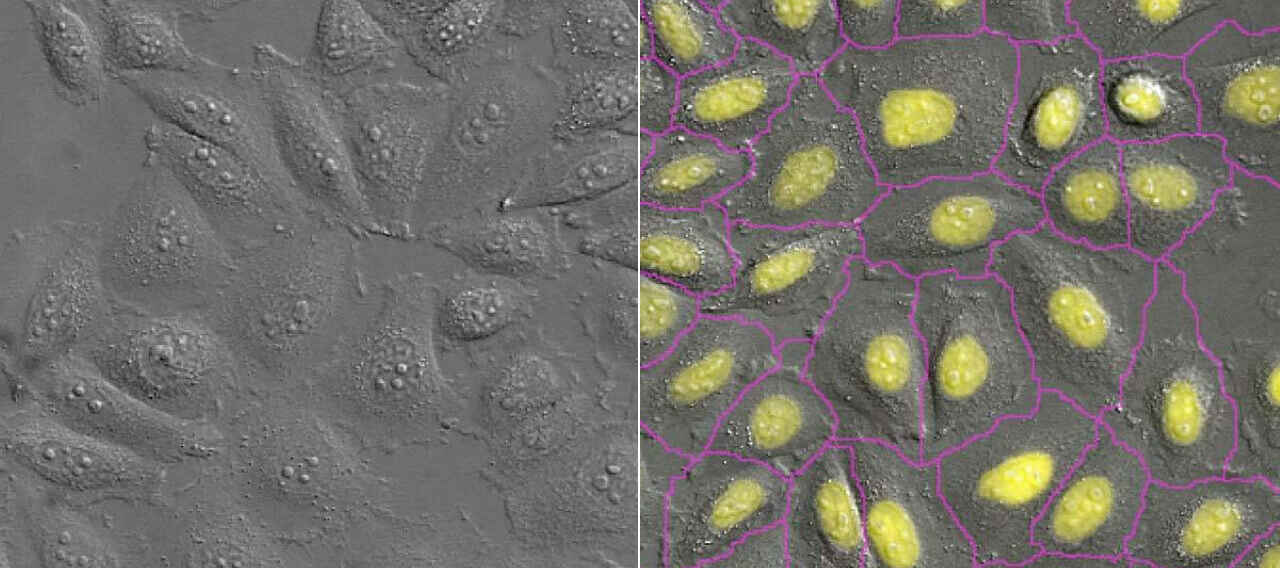
As an example of an appropriate use case, the Convert.ai module can be trained to predict DAPI nuclear staining patterns from transmitted light images. This predicted DAPI channel is useful and appropriate for cell counting, which depends mostly on the correct localization of DAPI signal. However, using predicted DAPI images for applications requiring quantification of intensity information would be inappropriate. While DL is reliable for accurately predicting DAPI localization and gross staining patterns, it is not a good choice for quantifying fine details and rare events, the latter of which would not be well represented in the training data to begin with and likely to be missed.
Streamlining Imaging and Analysis with Deep Learning
Image of a fluorescently labeled mouse intestine section acquired using a Nikon AX R resonant-scanning confocal microscope and 20X WI 0.95 NA objective lens
An advantage of DL methods is how quickly they can perform different and complex tasks. This allows for stacking of DL methods to perform complicated multi-part analyses in relatively short periods of time, even during the imaging process when time is limited.
For example, the Autosignal.ai module automatically determines the optimal laser power and gain within a matter of seconds when using our AX / AX R confocal microscope. A classical approach to this problem is unlikely to return a reliable solution in such a short period. Even a handful of seconds can make a significant difference when competing with factors such as photobleaching.
The Denoise.ai module is similarly fast, providing near-real time denoising. This makes for a synergistic combination with Autosignal.ai – the imaging conditions can be optimized within seconds with the push of a button. Other AI modules can be similarly stacked, such as Segment.ai for fast segmentation of image features, as shown in the video.
NIS.ai modules can be plugged into existing imaging and analysis pipelines within our NIS-Elements software. This is beneficial for optimizing the efficiency of high content-type experiments. The General Analysis 3 (GA3) module is used to craft customized measurement routines and allows users to apply NIS.ai DL-based and classical analyses together in the same pipeline. NIS.ai modules can also be embedded within the imaging routine using the JOBS experiment wizard – results obtained using an NIS.ai module within JOBS can be used to direct the course of the experiment. For example, one can automatically perform targeted acquisitions based on analysis results – a fitting choice for targeting rare events.
Deep Learning for Live Cell Imaging
Raw versus denoise resonant A1 timelapse
Drosophila sp. larvae sample provided by Amicia Elliot, PhD, NIH/NIMH, Bethesda MD. Acquired at the Quantitative Fluorescence Microscopy Course 2019.
Acquisition details:
Resonant scanner, Dimension 1024 x 512, Scan speed 30fps, Scan zoom x0.72, Averaging None
Objective Plan Apo 4X
One of the greatest benefits provided by many of the NIS.ai AI modules is the reduction of photon dose. Denoise.ai and Clarify.ai provide clearer images without requiring additional signal to be collected. Convert.ai allows for prediction of fluorescence imaging channels from transmitted light images (e.g., DIC), removing the need for fluorescence imaging altogether in some cases. Enhance.ai can predict higher signal images from low-signal data, minimizing the photon dose required for excitation.
Fluorescence microscopy generally requires much higher illumination intensity than transmitted light methods, has a limited photon budget, and is often the method of choice for live cell imaging, especially in research. Reduction of photon dose is useful for most imaging applications, but is essential for live cell fluorescence imaging, where photobleaching and phototoxicity are top concerns.
The Direction of AI and Deep Learning in Microscopy
Training NIS.ai modules is straightforward. The software provides a real-time view of training progress (Enhance.ai shown).
Analysis of low-signal DAPI image data (left) using the Enhance.ai module to predict a higher-signal restoration (right)
The application of AI and deep learning in microscopy is still in a relatively early stage, with new research and applications consistently being published and pushing the field forward. Given the potential of DL methods, and how quickly such methods are coming into the mainstream, it is important that the AI and microscopy communities develop clear guidelines for appropriate applications and quality metrics that allow non-specialists without computational backgrounds to be confident in using these tools for peer-reviewed research.
Nikon’s pre-trained AI modules, which currently includes Clarify.ai and Denoise.ai, do not allow for manual adjustment of ambiguous settings such as analysis “strength,” avoiding an opportunity for user bias to affect the results. Everyone who uses Clarify.ai or Denoise.ai is utilizing the same pre-trained network as every other user.
For NIS.ai modules that must be trained, including Convert.ai, Enhance.ai, and Segment.ai, the module includes an intuitive graphic user interface for network training. The user can monitor the progress of training, including a real-time view of the number of iterations and training loss.
Glossary
- Compatible Nikon Systems
- Indicates which Nikon microscopes/systems are compatible. This assumes the system is configured to capture digital images.
- Function
- Summary of the NIS.ai software module function.
- Pre-training Required
- Training of a deep learning neural network for image analysis is often performed using supervised learning with paired image data: an input image (of the sort that will be analyzed following training) and a ground truth image of the same area where the desired condition is met. The weights of the nodes within the neural network are adjusted to minimize the error between the predicted output image and the ground truth.
- Software Packages
- List of NIS-Elements software packages and modules available for purchase. Individual NIS.ai modules may be included or optionally available for the different software packages and modules as indicated.
- Home
- Applications
- Life Sciences
- Deep Learning in Microscopy