Dr. Steven Nedellec and Dr. Tiphaine Douanne
Dr. Steven Nedellec, Facility Manager of MicroPICell, Université de Nantes, France and Dr. Tiphaine Douanne, Universite de Nantes, Signaling in Oncogenesis, Angiogenesis and Permeability, CRCINA INSERM U1232, France
Nikon-Ausrüstung
- Eclipse Ti-E Inverted Microscope (see current model)
- A1 Confocal (see current model)
- N-SIM Super-Resolution
Dr. Steven Nedellec, Facility Manager of MicroPICell, Université de Nantes, France

What Nikon imaging technologies do you have in your facility?
We have four microscopes built by Nikon at our facility.
The first one we bought was the A1 confocal with spectral and resonant systems on it. The system was also installed with a Picoquant FLIM measuring system with a pulsed laser and a SPAD detector on it. A few years after, the system was upgraded with the resonant 1K scanning system. Recently, we had a machine learning plug-in dedicated to resonant denoising, also installed on the same imaging microscope.
We also installed, a few years after, another A1 classical inverted confocal microscope, on which also the super resolution SIM was installed – making it a multi modal fluorescence and super resolution microscopy imaging system.
Our facility is also developing what is called ‘micro patterning technology’, which was developed by the Alveole society based in France. And the system is also installed on the Nikon Ti Eclipse microscope. We use it to develop specific cell culture imaging.
The last one we installed was a quite classical inverted timelapse imaging but with a very powerful LED system for illumination with 4 channels. It was also installed with the JOBS module inside NIS software, which allows us to design quite complicated , quite rich experiments for high content screening imaging.
We are particularly interested in knowing how the A1 and N-SIM together has benefitted your users. Can you tell us about it, please?
First, we were very much impressed by the quality and the resolution with the super resolution SIM system. It provides great images, great resolution and also very strong contrast in the resulting image. For our users we were quite happy to see that manipulating the SIM microscope was quite easy. It is not so complicated to handle the software with different parameters. So as a facility manager, it’s quite a pleasure to have access to an easy-to-use advanced system, which is not always the case.
Moreover, the possibility of using multi channel, multi staining for super resolution is powerful for our users. Most of them are not microscopy specialists and they like to use the classical staining like they do for confocal and widefield microscopy. So SIM is very helpful for them.
Concerning the multimodality – confocal and SIM microscopy, it is also very powerful and very useful. It has a lot of advantages. For example, we have some scientific projects that need the multiscale imaging, by using the macro resolution confocal system, super resolution using SIM. Because it is on the same microscope, we can easily register and align these different images to put them side by side and have exhaustive information.

We are particularly interested in knowing how the A1 and N-SIM together has benefitted your users. Can you tell us about it, please?
First, we were very much impressed by the quality and the resolution with the super resolution SIM system. It provides great images, great resolution and also very strong contrast in the resulting image. For our users we were quite happy to see that manipulating the SIM microscope was quite easy. It is not so complicated to handle the software with different parameters. So as a facility manager, it’s quite a pleasure to have access to an easy-to-use advanced system, which is not always the case.
Moreover, the possibility of using multi channel, multi staining for super resolution is powerful for our users. Most of them are not microscopy specialists and they like to use the classical staining like they do for confocal and widefield microscopy. So SIM is very helpful for them.
Concerning the multimodality – confocal and SIM microscopy, it is also very powerful and very useful. It has a lot of advantages. For example, we have some scientific projects that need the multiscale imaging, by using the macro resolution confocal system, super resolution using SIM. Because it is on the same microscope, we can easily register and align these different images to put them side by side and have exhaustive information.
Can you tell us about your experience with Nikon microscopy systems?
My experience with Nikon microscope systems is that Nikon develops quite powerful and reliable microscopes. There are a lot of upgrades available. Even old systems can be upgraded, on site. We have access to quite new technologies even if the microscope was bought a few years ago. It is a very nice argument.
Moreover, Nikon also delivers a very powerful software, which of course can drive the microscope easily, but also gives us access to a lot of interesting options, such as machine learning deconvolution, machine learning denoising. The JOBS system is also very interesting because it allows to design quite rich, complicated experiments, which were not so easy to do a few years ago when JOBS was not available.
And lastly, I have to say that the local support of Nikon is very efficient. We have access to a lot of information and a lot of upgrades, and everything that we need.
Can you tell us about your experience with Nikon microscopy systems?
My experience with Nikon microscope systems is that Nikon develops quite powerful and reliable microscopes. There are a lot of upgrades available. Even old systems can be upgraded, on site. We have access to quite new technologies even if the microscope was bought a few years ago. It is a very nice argument.
Moreover, Nikon also delivers a very powerful software, which of course can drive the microscope easily, but also gives us access to a lot of interesting options, such as machine learning deconvolution, machine learning denoising. The JOBS system is also very interesting because it allows to design quite rich, complicated experiments, which were not so easy to do a few years ago when JOBS was not available.
And lastly, I have to say that the local support of Nikon is very efficient. We have access to a lot of information and a lot of upgrades, and everything that we need.
What challenges do researchers currently face and how do you think they can be overcome?
I think an important challenge, as a facility manager, is not mainly about optical improvement or improvement of the microscope by itself, but about the image processing. Such as what Nikon did by creating the machine learning denoising plug-in, and also by developing the JOBS module in NIS. And I also think that image compression can be an important challenge, because we are generating a lot of data. Through the images, it is quite heavy to store, it is quite complicated to share, and also it can be challenging to analyse it perfectly given the fact that the data is very big and difficult to handle. So I think image compression is also a major challenge for now.
What challenges do researchers currently face and how do you think they can be overcome?
I think an important challenge, as a facility manager, is not mainly about optical improvement or improvement of the microscope by itself, but about the image processing. Such as what Nikon did by creating the machine learning denoising plug-in, and also by developing the JOBS module in NIS. And I also think that image compression can be an important challenge, because we are generating a lot of data. Through the images, it is quite heavy to store, it is quite complicated to share, and also it can be challenging to analyse it perfectly given the fact that the data is very big and difficult to handle. So I think image compression is also a major challenge for now.
Dr. Tiphaine Douanne, Universite de Nantes, Signaling in Oncogenesis, Angiogenesis and Permeability, CRCINA INSERM U1232, France

Dr. Tiphaine Douanne, Universite de Nantes, Signaling in Oncogenesis, Angiogenesis and Permeability, CRCINA INSERM U1232, France
What do you image using A1 and N-SIM?
I use combined A1 and N-SIM to assess if a protein is involved in intracellular trafficking. For this we use HeLa cells and we stain them for CD63, which is a tetraspanin that is enriched in endosomes, lysosomes and intracellular vesicles. We also use GM130, which is a Golgi apparatus staining, in order to orientate within our cell. The idea is to silence different proteins of interest by RNA interference, and to assess if they have any effects on the staining.
What do you image using A1 and N-SIM?
I use combined A1 and N-SIM to assess if a protein is involved in intracellular trafficking. For this we use HeLa cells and we stain them for CD63, which is a tetraspanin that is enriched in endosomes, lysosomes and intracellular vesicles. We also use GM130, which is a Golgi apparatus staining, in order to orientate within our cell. The idea is to silence different proteins of interest by RNA interference, and to assess if they have any effects on the staining.
Why do you use A1 and N-SIM?
We use both A1 and N-SIM together because they are quite complementary techniques. Early on, observations with A1 allowed us to see that removing some proteins had an effect on our staining. It drove the staining to polarise within the cell when normally it is quite diffused. So this allowed us to quantify the phenotype on a polarised vs non-polarised basis.
Then to get a closer look at the structure we moved on to N-SIM. And we could see for instance that some structures that appeared as big aggregates by confocal turned out to be smaller vesicles clumped together by N-SIM.
Why do you use A1 and N-SIM?
We use both A1 and N-SIM together because they are quite complementary techniques. Early on, observations with A1 allowed us to see that removing some proteins had an effect on our staining. It drove the staining to polarise within the cell when normally it is quite diffused. So this allowed us to quantify the phenotype on a polarised vs non-polarised basis.
Then to get a closer look at the structure we moved on to N-SIM. And we could see for instance that some structures that appeared as big aggregates by confocal turned out to be smaller vesicles clumped together by N-SIM.
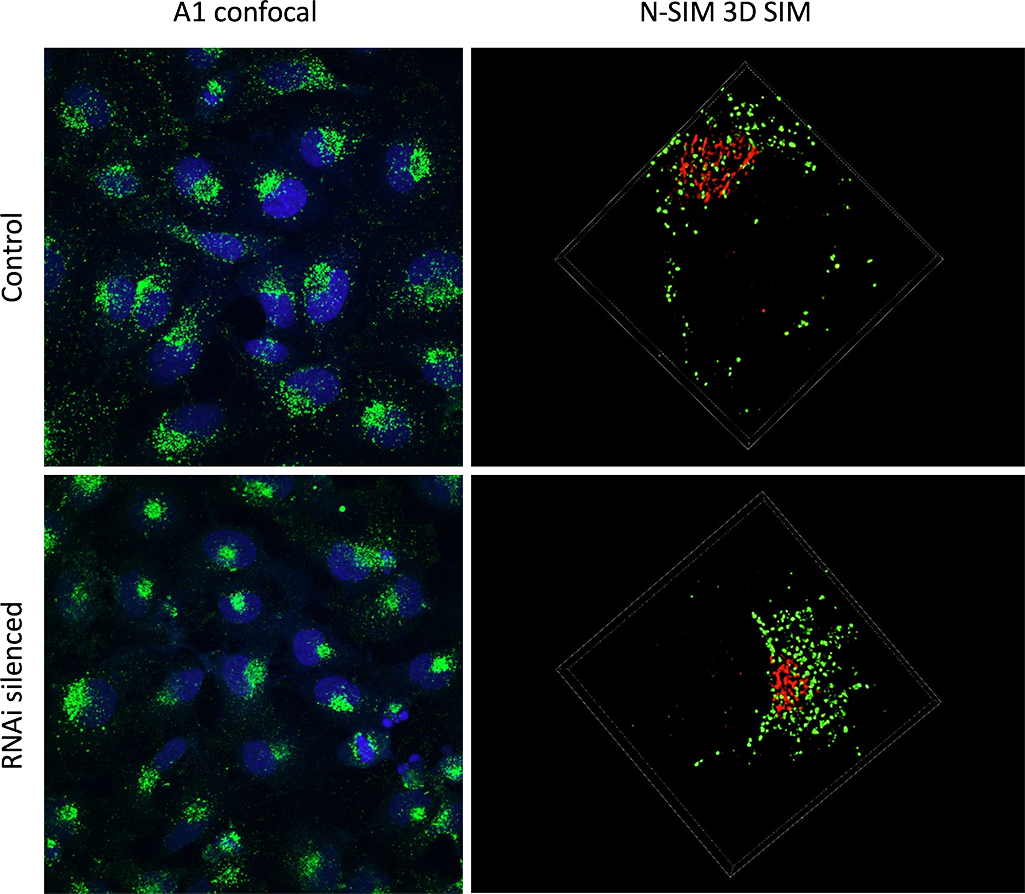
The RNAi treated HeLa cells display a polarized phenotype.
A1 confocal imaging was used to identify polarized cells vs diffused cells within the population (left column). 3D SIM imaging was applied to specific cells to discern the small structural details (right column). The SIM resolution enables the quantification of individual vesicles (green), and the study of their distribution in relation to the Golgi apparatus (red).
Is it important for you to have both A1 and N-SIM on the same platform? And if so, why is that?
It is important for us to have both A1 and N-SIM on the same platform because the techniques nicely complement each other.
The original confocal observation allowed us to have a general view and to screen for more proteins. Once we found phenotypes of interest we moved onto the N-SIM to have a more discriminative view.
On a more practical note, because polarization is very important in our phenotype, it was interesting to be able to orientate the cells using the confocal image and then move onto the SIM to take a Z-stack, which could be a bit more time consuming.
Is it important for you to have both A1 and N-SIM on the same platform? And if so, why is that?
It is important for us to have both A1 and N-SIM on the same platform because the techniques nicely complement each other.
The original confocal observation allowed us to have a general view and to screen for more proteins. Once we found phenotypes of interest we moved onto the N-SIM to have a more discriminative view.
On a more practical note, because polarization is very important in our phenotype, it was interesting to be able to orientate the cells using the confocal image and then move onto the SIM to take a Z-stack, which could be a bit more time consuming.
What have A1 and N-SIM together enabled you to find out, which would otherwise be difficult to achieve?
Having A1 and N-SIM together enabled us to identify proteins involved in intracellular trafficking. We were able to quantify the size and the number of structures ranging from 40-100 nm3 and this was instrumental in determining if a protein is involved in endosomal trafficking.
What have A1 and N-SIM together enabled you to find out, which would otherwise be difficult to achieve?
Having A1 and N-SIM together enabled us to identify proteins involved in intracellular trafficking. We were able to quantify the size and the number of structures ranging from 40-100 nm3 and this was instrumental in determining if a protein is involved in endosomal trafficking.
