- fr Change Region
- Global Site
- Accueil
- Ressources
- Notes d'application
Notes d'application
High-sensitivity confocal microscope detector NSPARC enables live imaging of autophagosomes.
novembre 2024
Autophagy is a critical degradation pathway in eukaryotic cells, essential for maintaining cellular homeostasis. Dysfunction in this process has been implicated in the onset and progression of cancer and neurodegenerative diseases. Upon induction of autophagy, a flattened membrane structure known as the isolation membrane emerges near the endoplasmic reticulum (ER), leading to the formation of double-membrane vesicles approximately 1 μm in diameter called autophagosomes that sequester cytoplasmic material. In mammalian cells, numerous autophagosomes form on the ER within an hour before eventually fusing with lysosomes, where their contents are degraded and nutrients are recycled. Despite extensive study, the detailed mechanisms of autophagosome formation remain unclear.
This application note presents an example of the dynamic membrane deformation process involved in autophagosome formation, observed using the super-resolution confocal laser-scanning microscope system AX with NSPARC. The NSPARC detector achieves higher resolution and signal-to-noise ratio compared to traditional confocal microscopy. This improved image quality can be leveraged to conduct faster live-cell imaging with lower laser power while still capturing subcellular detail. With NSPARC, three-dimensional visualization of the autophagosome formation process is possible with reduced cellular damage from laser exposure. Using NSPARC, researchers were able to capture the state of the autophagosome opening just before closure and the movement of the isolation membrane in three dimensions.
Keywords: Live imaging, autophagosome, isolation membrane
Research background
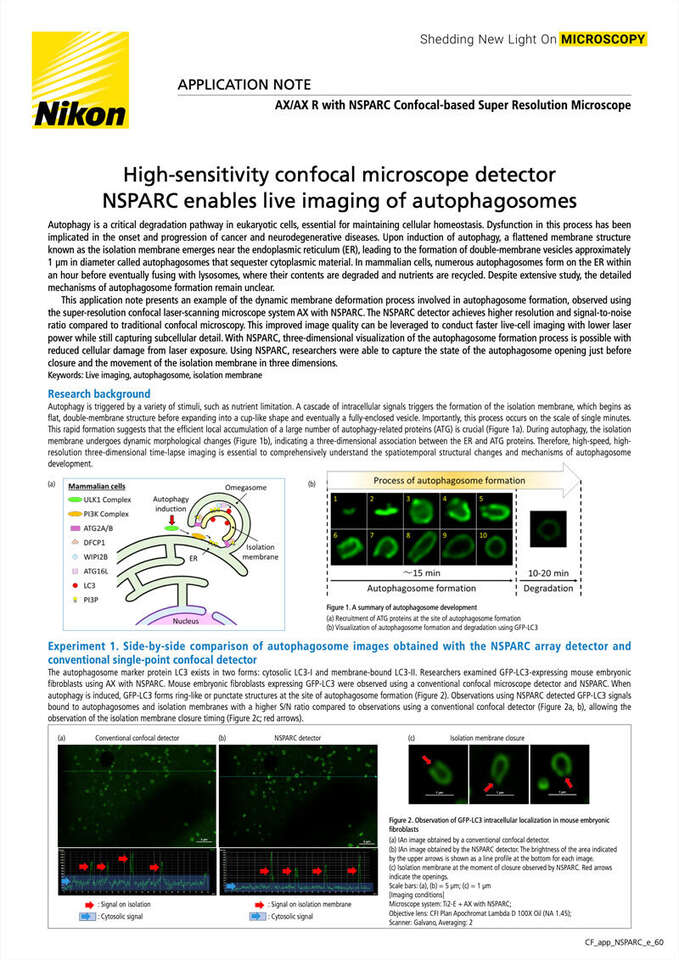
Autophagy is triggered by a variety of stimuli, such as nutrient limitation. A cascade of intracellular signals triggers the formation of the isolation membrane, which begins as flat, double-membrane structure before expanding into a cup-like shape and eventually a fully-enclosed vesicle. Importantly, this process occurs on the scale of single minutes. This rapid formation suggests that the efficient local accumulation of a large number of autophagy-related proteins (ATG) is crucial (Figure 1a). During autophagy, the isolation membrane undergoes dynamic morphological changes (Figure 1b), indicating a three-dimensional association between the ER and ATG proteins. Therefore, high-speed, high-resolution three-dimensional time-lapse imaging is essential to comprehensively understand the spatiotemporal structural changes and mechanisms of autophagosome development.
Figure 1. A summary of autophagosome development
(a) Recruitment of ATG proteins at the site of autophagosome formation
(b) Visualization of autophagosome formation and degradation using GFP-LC3
Experiment 1. Side-by-side comparison of autophagosome images obtained with the NSPARC array detector and conventional single-point confocal detector
The autophagosome marker protein LC3 exists in two forms: cytosolic LC3-I and membrane-bound LC3-II. Researchers examined GFP-LC3-expressing mouse embryonic fibroblasts using AX with NSPARC. Mouse embryonic fibroblasts expressing GFP-LC3 were observed using a conventional confocal microscope detector and NSPARC. When autophagy is induced, GFP-LC3 forms ring-like or punctate structures at the site of autophagosome formation (Figure 2). Observations using NSPARC detected GFP-LC3 signals bound to autophagosomes and isolation membranes with a higher S/N ratio compared to observations using a conventional confocal detector (Figure 2a, b), allowing the observation of the isolation membrane closure timing (Figure 2c; red arrows).
Figure 2. Observation of GFP-LC3 intracellular localization in mouse embryonic fibroblasts
(a) An image obtained by a conventional confocal detector.
(b) An image obtained by the NSPARC detector. The brightness of the area indicated by the upper arrows is shown as a line profile at the bottom for each image.
(c) Isolation membrane at the moment of closure observed by NSPARC. Red arrows indicate the openings.
Scale bars: (a), (b) = 5 µm; (c) = 1 µm
[Imaging conditions]
Microscope system: ECLIPSE Ti2-E + AX with NSPARC
Objective lens: CFI Plan Apochromat Lambda D 100X Oil (NA 1.45)
Scanner: Galvano, Averaging: 2
Figure 3. Live, three-dimensional imaging of autophagosome formation
Mouse embryonic fibroblasts expressing GFP-LC3 were observed using AX with NSPARC. The captured fluorescence images were deconvolved using the Richardson-Lucy method.
(a) 3D view of autophagosomes forming over 9 minutes. The arrow indicates a single autophagosome. Dynamic changes in the closure of the isolation membrane were captured between 4 and 8 minutes (yellow arrow). Tick marks indicate position in µm.
(b) To capture the movement of the autophagosome in the Z-axis direction, the XY plane images of a single autophagosome in the obtained images were arranged along the time axis in the Z-axis direction. There was significant movement in the Z direction during the 200-450 second timing when isolation membrane formation occurs.
[Imaging conditions]
Microscope system: ECLIPSE Ti2-E + AX with NSPARC
Objective lens: CFI Plan Apochromat Lambda D 100X Oil (NA 1.45)
Scanner: Galvano, Averaging: None
Z-step: 0.15 µm, Z acquisition range: 2 µm
Experiment 2: Live, high-speed, three-dimensional imaging of autophagosome formation using NSPARC
To visualize the dynamic morphological changes during autophagosome formation in detail, researchers performed live, high-speed imaging using AX with NSPARC, acquiring sectioned images over a 2 μm volume. Despite capturing numerous images were along the Z-axis at short intervals for optimal deconvolution results, rapid data acquisition was achieved with a time of approximately 1.5 seconds per stack. Even during long-term imaging at 10-second intervals, photobleaching and cellular damage were minimal. Moreover, the acquired images had a high enough S/N ratio to observe the state of the autophagosome opening just before closure (Figure 3a). This rapid, volumetric imaging revealed that the isolation membrane moves significantly in all three dimensions during autophagosome formation (Figure 3b).
Conclusions
Experiments in this application note have revealed that the isolation membrane moves dynamically in three-dimensional space during autophagosome formation. To understand the behavior of associated proteins, it is crucial to acquire data with both high signal-to-noise (S/N) ratio and rapid speed. Quantification of fluorescence signals is commonly used as an indicator when screening autophagy-regulating drugs. By utilizing AX R with NSPARC, which enables high temporal resolution observations while maintaining sufficient S/N ratio, researchers achieved more accurate data acquisition compared to conventional confocal microscopy. Moreover, the accelerated data acquisition minimizes laser-induced sample damage and fluorescence photobleaching. In the future, we anticipate that this technology will enable long-term observations of autophagosome formation processes and the effects of autophagy-regulating drugs, which is challenging with conventional confocal microscopes.
Acknowledgment
We would like to express our sincere gratitude to Dr. Nobuo Noda and Dr. Yuta Ogasawara of Institute for Genetic Medicine, Hokkaido University for their great cooperation and generous advice.
Website for Biological Molecular Mechanisms lab:
https://mechanism.igm.hokudai.ac.jp/
Product information
AX/AX R with NSPARC Confocal-based Super Resolution Microscope
The super-resolution detector NSPARC features an array of 25 subunit detectors to achieve higher resolution and signal to noise ratio without comprising the functionality of the conventional AX/AX R confocal microscope.
- Accueil
- Ressources
- Notes d'application
