- fr Change Region
- Global Site
- Accueil
- Produits
- Microscopes confocaux et multiphotoniques
- AX R MP with NSPARC
AX R MP with NSPARC
Système de microscope multiphotonique
Software Powered by Artificial Intelligence
Software for deep, wide imaging
NIS-Elements C control software enables centralization of workflow from image acquisition to analysis, making it easy to customize experiment templates that combine multiple settings.
AI software innovations designed to assist
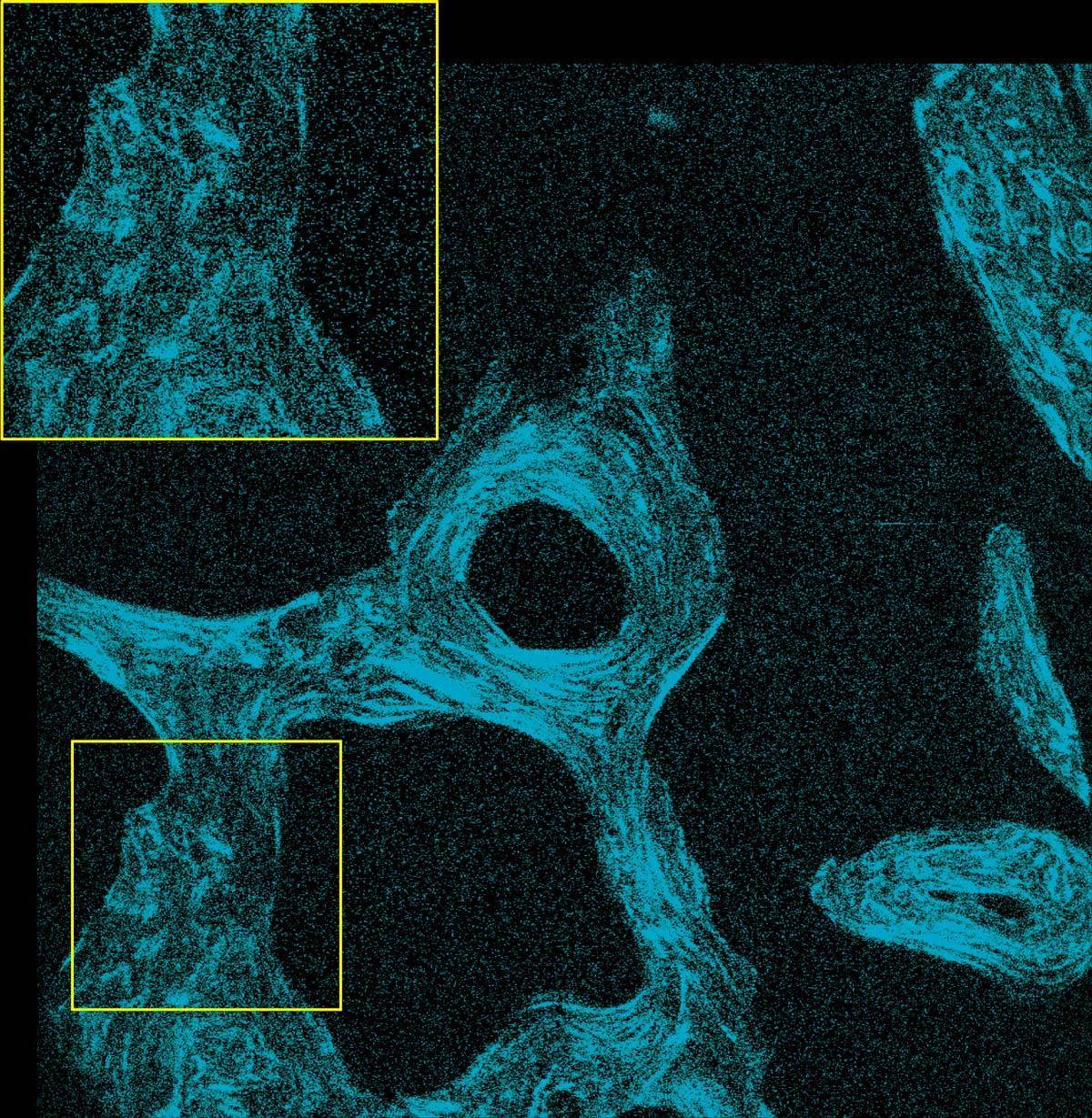
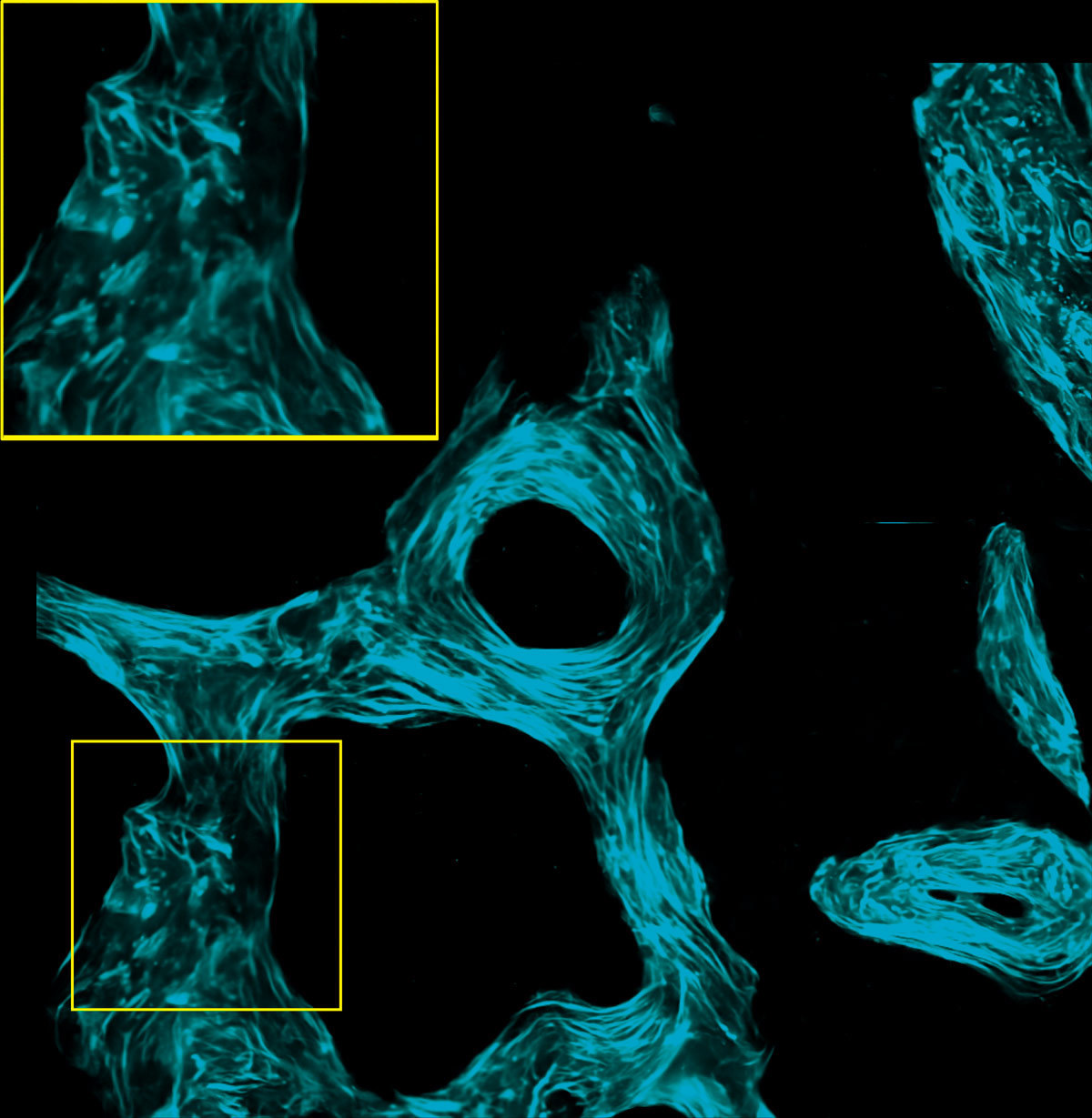
SHG images of un-decalcified bone section of a monkey captured at 920 nm IR excitation wavelength
Image courtesy of Dr. Tadahiro Iimura and Dr. Takanori Sato of the Department of Pharmacology, Faculty and Graduate School of Dental Medicine, Hokkaido University
The optional software module NIS.ai is equipped with image processing tools and customization functions. Utilizing deep learning and AI technology, it automates image acquisition and generation of optimal images for analysis. With NIS.ai, users can build in tailor-made solutions for acquisition, visualization and analysis customized for specific experiments.
Denoise.ai, a standard module in the NIS-Elements C imaging software, automatically removes Poisson shot noise from resonant confocal images. Resonant scanning results in ultrashort (tens of nanoseconds) dwell times that are extremely favorable for reducing phototoxicity and increasing specimen viability for long term imaging. While resonant scanning at very short exposure times usually requires line averaging to reduce Poisson shot noise contributions, users instead can employ Denoise.ai to eliminate the noise component. Denoise.ai can recognize and remove the shot noise components of images, increasing clarity and allowing for shorter exposure times and longer time-lapse experiments, while maintaining viability.
Unmix wavelength crosstalk
When there is significant crosstalk between multiple wavelengths excited by one wavelength, fluorescent separation (unmixing) allows clear separation of dyes.
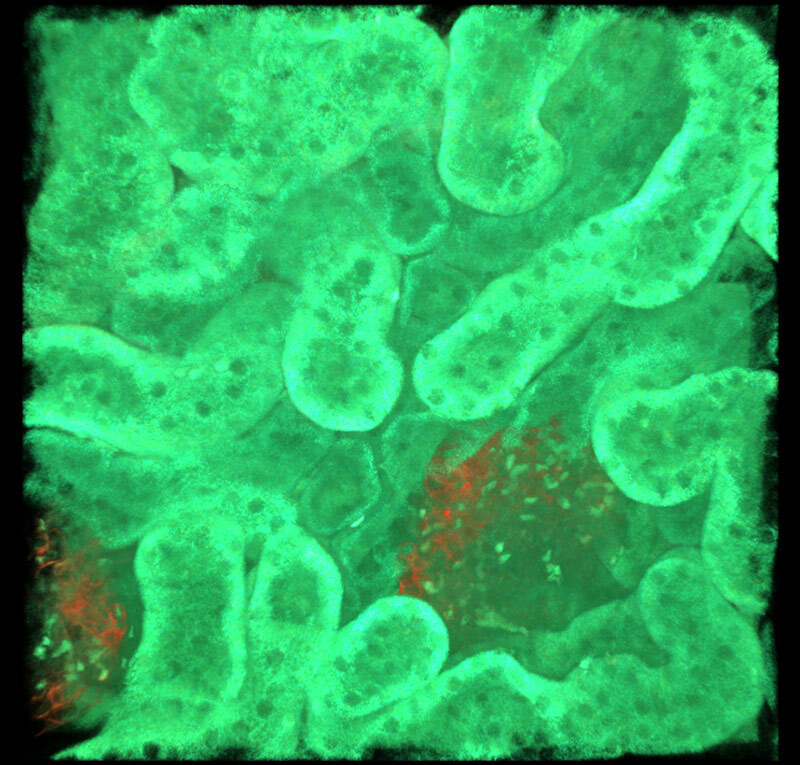
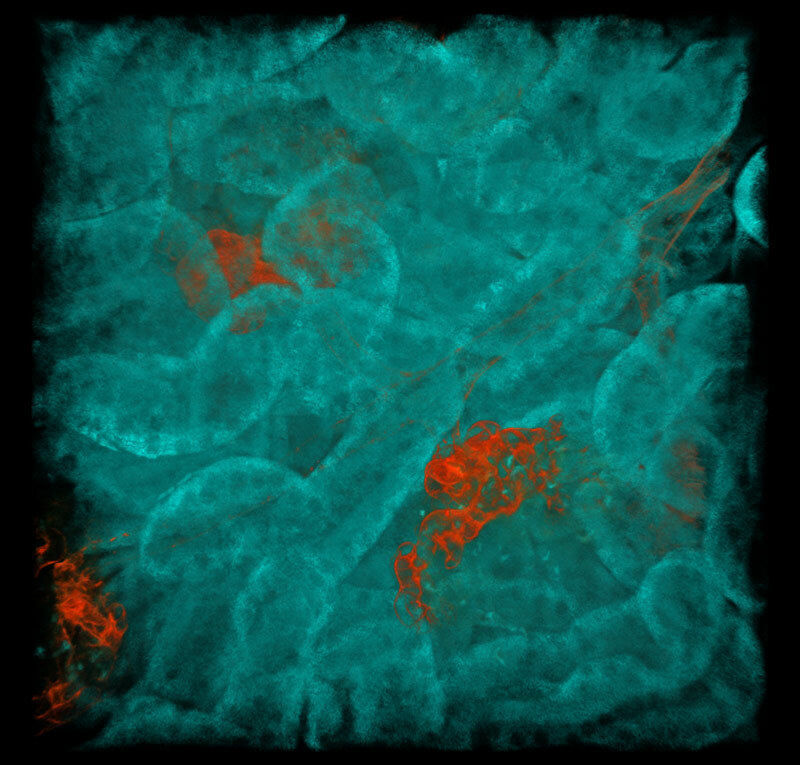
All channels merged
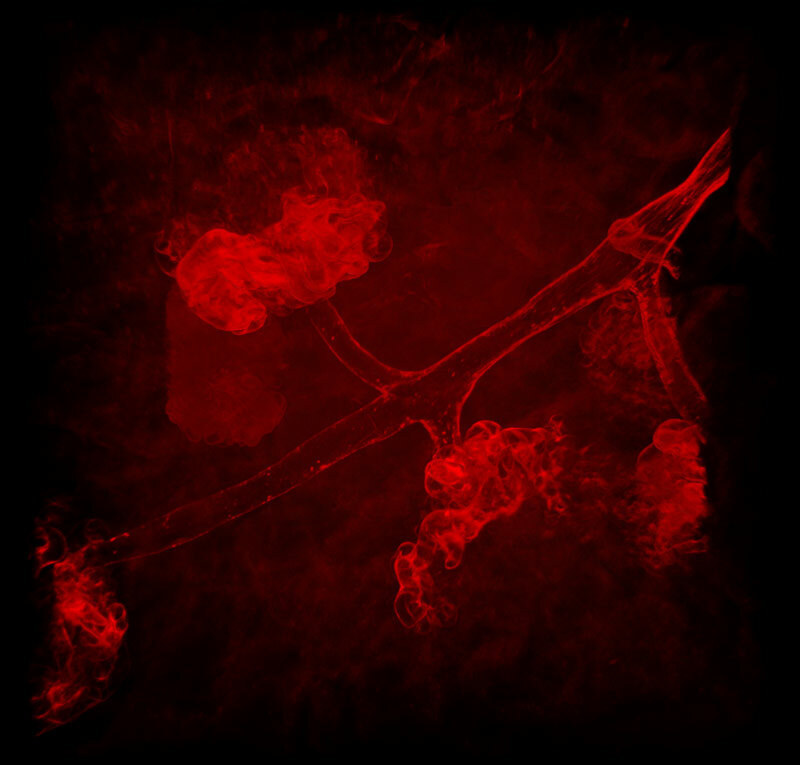

Red only
Multicolor fluorescent images of mouse kidney
Red: blood vessels (Alexa Fluor 594), Cyan: SHG, Green: Autofluorescence
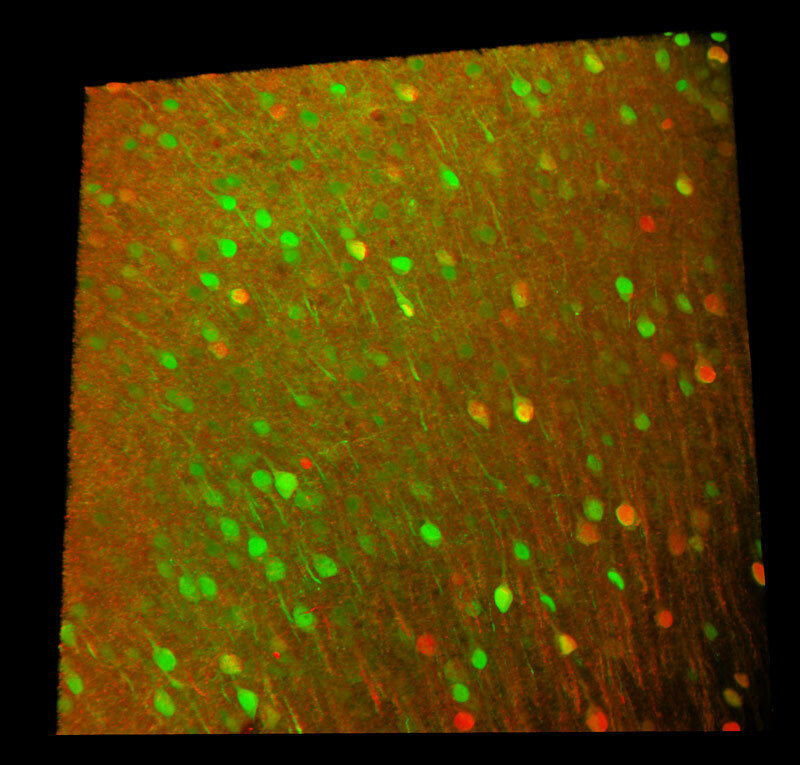
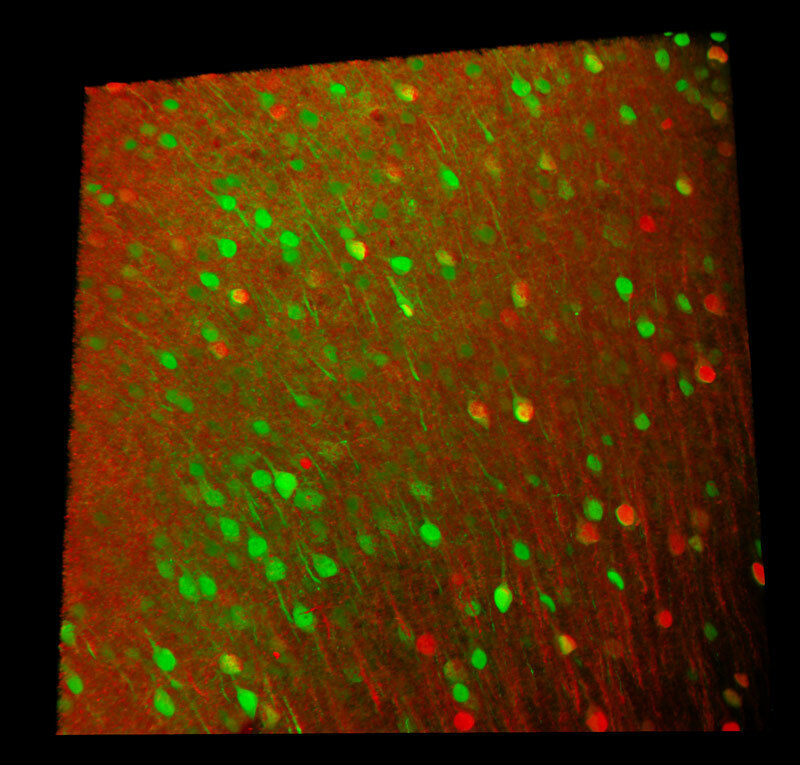
All channels merged
Two AAV– (AAV-Camk2-Cre and AAV-hSyn-GFP) were injected into the cerebral cortex of an Ai14 mouse to express GFP and TdTomato in neurons.
Red: AAV-Camk2-Cre neurons (TdTomato)
Green: AAV-hSyn-GFP neurons (GFP)
Image courtesy of Dr. Aya Ishida, RIKEN Center for Brain Science, Laboratory for Brain Development and Disorders
Deconvolution improves image quality in deep areas
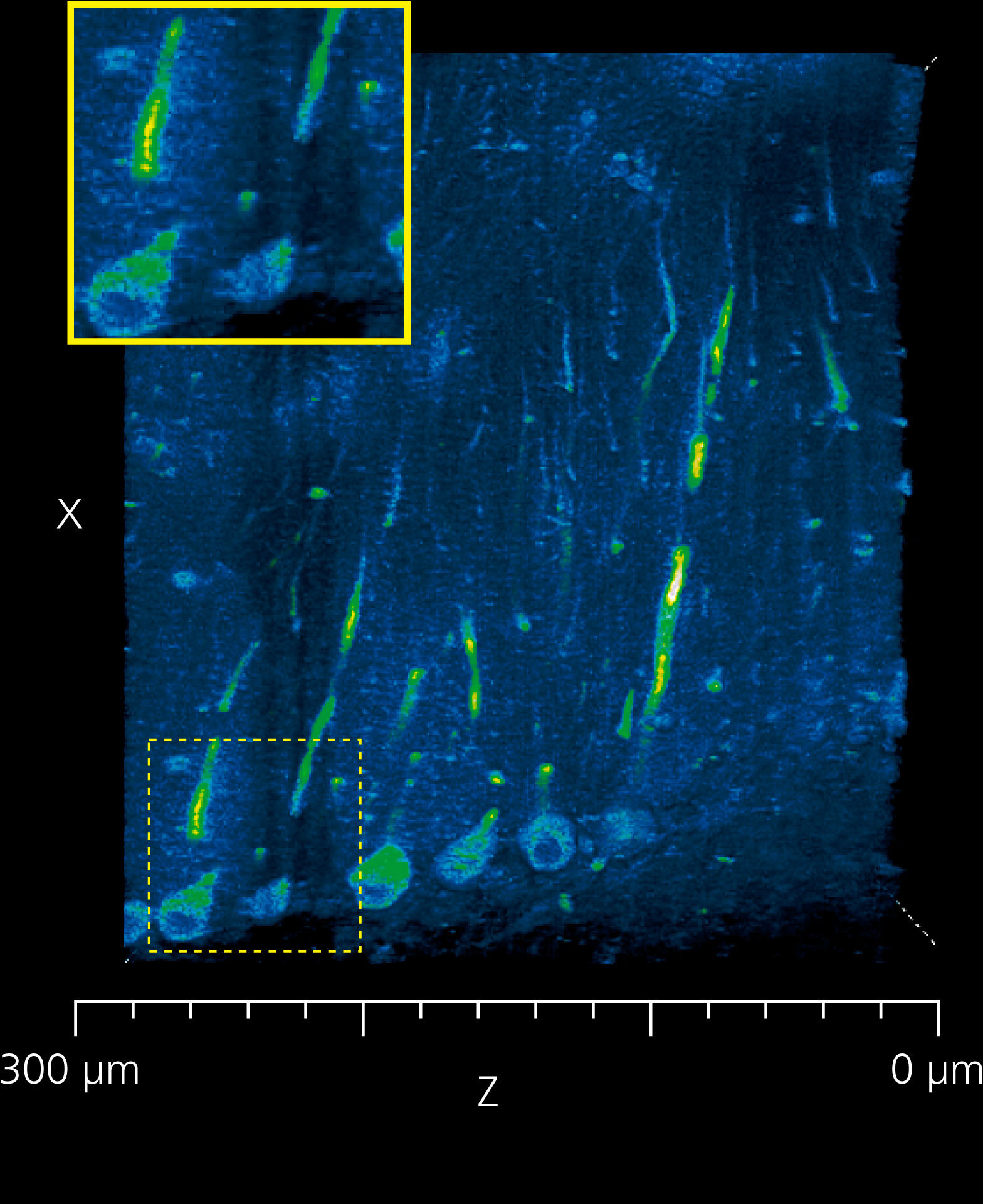
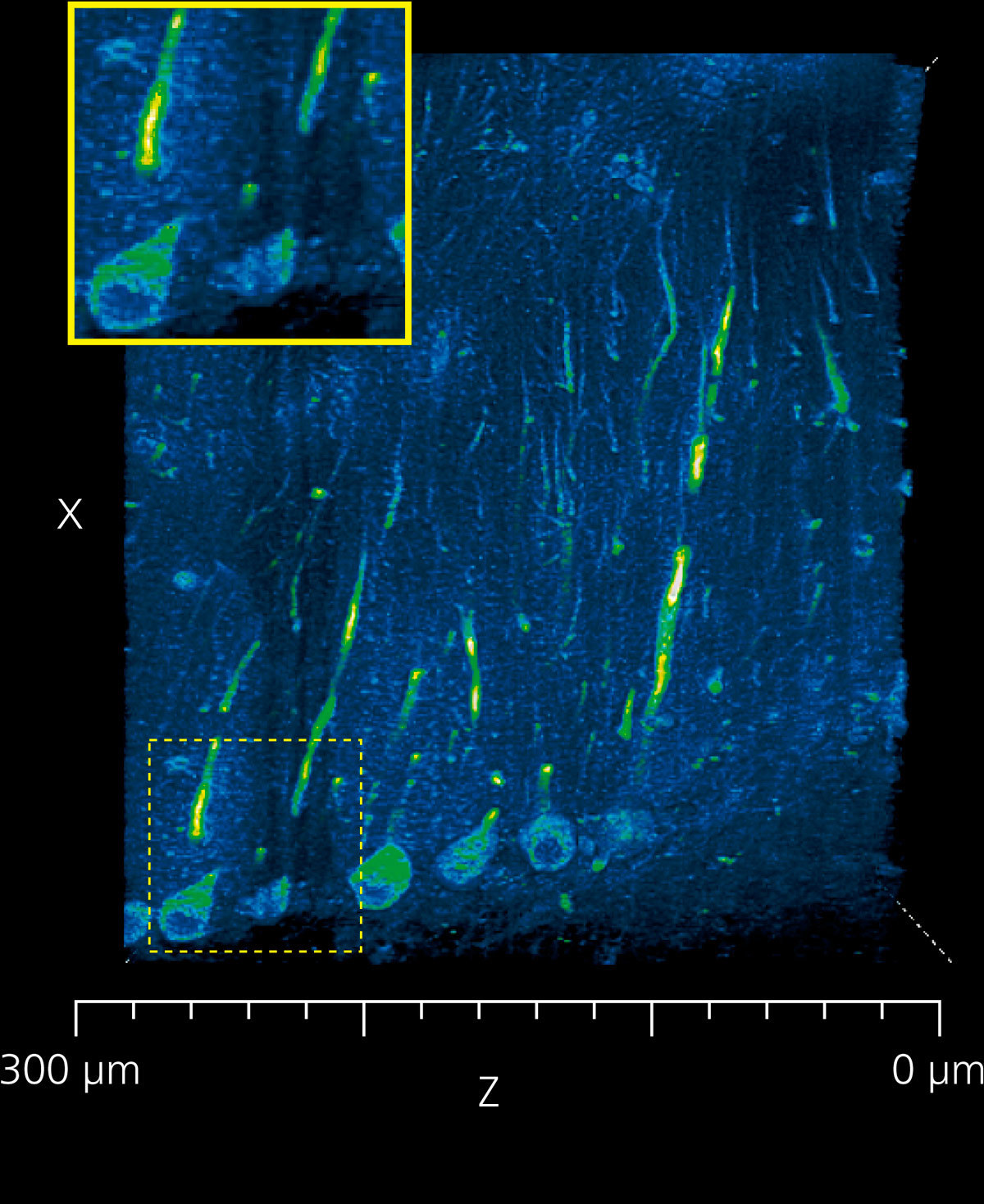
Deconvolution processing is effective in capturing detailed structures in deep areas because it reduces image elongation in the optical axis direction.
Slices of the cerebellum of a LC3GFP mouse.
The right side of each image is the surface layer and left side is at a depth of about 300 μm.
Blue: cerebellum (autofluorescence)
Green: Purkinje cells
Image courtesy of Dr. Laurence Dubreil, Dr. Julien Pichon and Pr. Marie-Anne Colle, PAnTher UMR703 INRAE/ Oniris, Nantes France
Flat field of view
Nikon's core strength is optical design. This requires that all aspects of our objectives and systems be mathematically calculated. With this information, NIS-Elements can predict “vignetting” or "shading" across a given field and correct for it automatically. With the click of a button, new “AX Shading Correction" provides flat images across maximized fields of view.
Seamlessly stitched images
When a sample is too large for a single field of view, stitching is the tool of choice. With ”AX Shading Correction,“ flat images generate seamlessly stitched data; reducing or eliminating common lines and artifacts.

