- es Change Region
- Global Site
Notas de aplicación
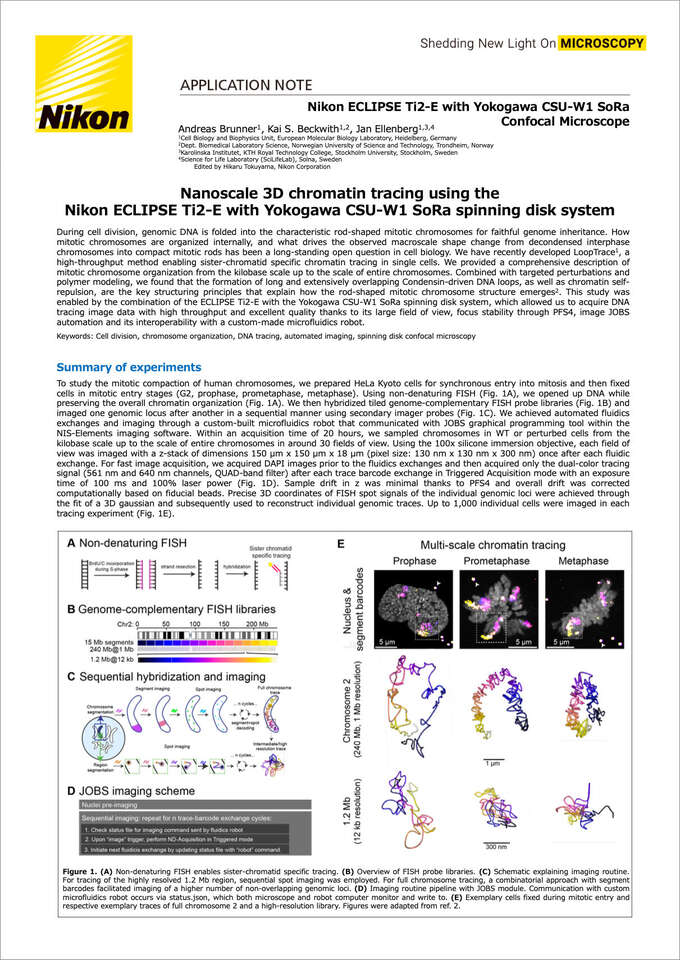
Nanoscale 3D chromatin tracing using the Nikon ECLIPSE Ti2-E with Yokogawa CSU-W1 SoRa spinning disk system
diciembre 2025
During cell division, genomic DNA is folded into the characteristic rod-shaped mitotic chromosomes for faithful genome inheritance. How mitotic chromosomes are organized internally, and what drives the observed macroscale shape change from decondensed interphase chromosomes into compact mitotic rods has been a long-standing open question in cell biology. We have recently developed LoopTrace1, a high-throughput method enabling sister-chromatid specific chromatin tracing in single cells. We provided a comprehensive description of mitotic chromosome organization from the kilobase scale up to the scale of entire chromosomes. Combined with targeted perturbations and polymer modeling, we found that the formation of long and extensively overlapping Condensin-driven DNA loops, as well as chromatin self-repulsion, are the key structuring principles that explain how the rod-shaped mitotic chromosome structure emerges2. This study was enabled by the combination of the ECLIPSE Ti2-E with the Yokogawa CSU-W1 SoRa spinning disk system, which allowed us to acquire DNA tracing image data with high throughput and excellent quality thanks to its large field of view, focus stability through PFS4, image JOBS automation and its interoperability with a custom-made microfluidics robot.
Keywords: Cell division, chromosome organization, DNA tracing, automated imaging, spinning disk confocal microscopy
References
- Beckwith, K.S., Ødegård-Fougner, Ø., Morero, N.R., Barton, C., Schueder, F., Tang, W., Alexander, S., Peters, J.-M., Jungmann, R., Birney, E., Ellenberg, J. (2025). Nanoscale 3D DNA tracing in non-denatured cells resolves the Cohesin-dependent loop architecture of the genome in situ. Nature Communications 16(1): 6673. https://doi.org/10.1038/s41467-025-61689-y
- Beckwith, K.S., Brunner, A., Morero, N.R., Jungmann, R., and Ellenberg, J. (2025). Nanoscale DNA tracing reveals the self-organization mechanism of mitotic chromosomes. Cell 188(10), 2656-2669.e17. https://doi.org/10.1016/J.CELL.2025.02.028
