- de Change Region
- Global Site
- Home
- intergrundwissen
- Application Notes
Application Notes
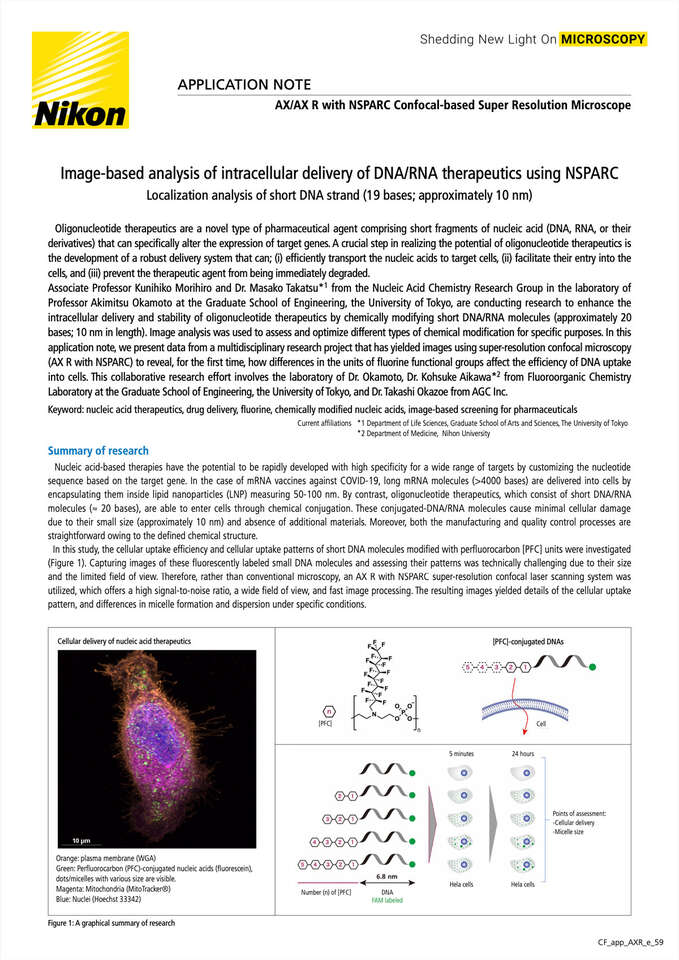
Image-based analysis of intracellular delivery of DNA/RNA therapeutics using NSPARC
Localization analysis of short DNA strand (19 bases; approximately 10 nm)
September 2024
Oligonucleotide therapeutics are a novel type of pharmaceutical agent comprising short fragments of nucleic acid (DNA, RNA, or their derivatives) that can specifically alter the expression of target genes. A crucial step in realizing the potential of oligonucleotide therapeutics is the development of a robust delivery system that can; (i) efficiently transport the nucleic acids to target cells, (ii) facilitate their entry into the cells, and (iii) prevent the therapeutic agent from being immediately degraded.
Associate Professor Kunihiko Morihiro and Dr. Masako Takatsu*1 from the Nucleic Acid Chemistry Research Group in the laboratory of Professor Akimitsu Okamoto at the Graduate School of Engineering, the University of Tokyo, are conducting research to enhance the intracellular delivery and stability of oligonucleotide therapeutics by chemically modifying short DNA/RNA molecules (approximately 20 bases; 10 nm in length). Image analysis was used to assess and optimize different types of chemical modification for specific purposes. In this application note, we present data from a multidisciplinary research project that has yielded images using super-resolution confocal microscopy (AX R with NSPARC) to reveal, for the first time, how differences in the units of fluorine functional groups affect the efficiency of DNA uptake into cells. This collaborative research effort involves the laboratory of Dr. Okamoto, Dr. Kohsuke Aikawa*2 from Fluoroorganic Chemistry Laboratory at the Graduate School of Engineering, the University of Tokyo, and Dr. Takashi Okazoe from AGC Inc.
Keywords: nucleic acid therapeutics, drug delivery, fluorine, chemically modified nucleic acids, image-based screening for pharmaceuticals
Current affiliations:
*1 Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo.
*2 Department of Medicine, Nihon University.
Summary of research
Nucleic acid-based therapies have the potential to be rapidly developed with high specificity for a wide range of targets by customizing the nucleotide sequence based on the target gene. In the case of mRNA vaccines against COVID-19, long mRNA molecules (>4000 bases) are delivered into cells by encapsulating them inside lipid nanoparticles (LNP) measuring 50-100 nm. By contrast, oligonucleotide therapeutics, which consist of short DNA/RNA molecules (≈ 20 bases), are able to enter cells through chemical conjugation. These conjugated-DNA/RNA molecules cause minimal cellular damage due to their small size (approximately 10 nm) and absence of additional materials. Moreover, both the manufacturing and quality control processes are straightforward owing to the defined chemical structure.
In this study, the cellular uptake efficiency and cellular uptake patterns of short DNA molecules modified withperfluorocarbon [PFC] units were investigated (Figure 1). Capturing images of these fluorescently labeled small DNA molecules and assessing their patterns was technically challenging due to their size and the limited field of view. Therefore, rather than conventional microscopy, an AX R with the NSPARC super-resolution confocal laser scanning system was utilized, which offers a high signal-to-noise ratio, a wide field of view, and fast image processing. The resulting images yielded details of the cellular uptake pattern, and differences in micelle formation and dispersion under specific conditions.
Cellular delivery of nucleic acid therapeutics
Orange: plasma membrane (WGA)
Green: Perfluorocarbon (PFC)-conjugated nucleic acids (fluorescein), dots/micelles with various size are visible.
Magenta: Mitochondria(MitoTracker®)
Blue: Nuclei (Hoechst 33342)
Figure 2: Observation of PFC-DNA conjugates introduced into HeLa cells.
(A) Images of cells 24 hours post-addition of PFC-DNA conjugates ([PFC]n-DNA, n = 2 or 4) acquired by A1 confocal microscopy or NSPARC. The images on the right side of each panel are magnified views of the white boxed areas in the corresponding images on the left side. Large aggregations detected by NSPARC in an image with [PFC]4-DNA were indicated by red arrows.
(B) NSPARC observation images of cells taken 5 minutes and 24 hours post-addition of 1.0 μM of [PFC]0,1,2,3,4,5-DNA. The images were acquired in the z-axis range of approximately 10 µm with an interval of 0.15 µm, and the maximum intensity projection (MIP) images are shown. The images on the right side of each panel are magnified views of the white boxed areas in the corresponding images on the left side.
White: Plasma membrane (WGA) - only in the left images
Green: Perfluorocarbon (PFC)-conjugated nucleic acids
Blue: Nucleus (Hoechst 33342)
Scale bars: 10 µm
[Image acquisition conditions]
NSPARC Microscope System: Ti2-E+AX R with NSPARC
Objective: CFI Plan Apochromat Lambda D 100X Oil (NA 1.45)
Scanner: Galvano, Averaging: None
A1 Microscope System: Ti-E+A1
Objective: CFI Plan Apochromat VC 60X Oil (NA 1.4)
Scanner: Galvano, Averaging: 8 times
Adapted with permission from M. Takatsu, K. Morihiro, H. Watanabe, M. Yuki, T. Hattori, K. Noi, K. Aikawa, K. Noguchi, M. Yohda, T. Okazoe, and A. Okamoto, Cellular penetration and intracellular dynamics of perfluorocarbon-conjugated DNA/RNA as a potential means of conditional nucleic acid delivery, ACS Chem. Biol. 18(12), Nov 19, 2023, 2590-2598. https://doi.org/10.1021/acschembio.3c00612. Copyright 2023 American Chemical Society
Results
The cellular uptake of fluorescently labeled [PFC]-conjugated DNA was examined using confocal laser scanning microscopy. Two microscopy systems were initially tested: the AX R with NSPARC, which features a detector array capable of super-resolution imaging, and the A1 confocal microscope with a conventional single point detector (Figure 2A). HeLa cells were grown in culture medium and imaged after addition of the chemically modified nucleic acids ([PFC]n-DNA, where n denotes the number of [PFC] units). The difference in micelle size could not be distinguished using A1 conventional confocal microscopy. However, using the AX R with NSPARC, clear differences between monodisperse micelles ([PFC]2-DNA) and polydisperse micelles ([PFC]4-DNA) were observed (Figure 2A). Within five minutes after introducing the nucleic acids into the cells, [PFC]2-DNA exhibited an array of small dots, [PFC]3-DNA and [PFC]5-DNA generated monodisperse micelles, whereas [PFC]4-DNA formed polydisperse micelles on the cell membrane. At 24 hours post application, [PFC]3-DNA maintained its initial micelle state, while [PFC]5-DNA changed into a polydisperse micelle state (Figure 2B). Imaging time was reduced by approximately one-tenth, and the field of view was 3.8 times larger than other super-resolution microscopy techniques based on structured illumination. This allowed cell- and population-level trends to be captured within a reasonable time frame.Our observations showed that [PFC]2, with the least number of PFC-containing units, was efficiently taken up into the cells. Moreover, the introduction of [PFC]2-modified siRNA into cells caused gene-silencing effects (refer to the cited literature).
Conclusion and future perspectives
Using the AX R with NSPARC imaging system, intracellular delivery and stability of short [PFC]-conjugated DNA could be assessed by capturing high resolution images with a high signal-to-noise ratio. Slight differences in DNA modification changed the micelle formation and dispersion states of the DNA molecules. Furthermore, these states affected the efficiency of cellular uptake and intracellular stability. Based on the imaging data and biological activity, it was possible to analyze how chemical modification changes in the DNA molecules affect their functionality as oligonucleotide therapeutics. This study represents a significant step towards developing an image-based technology platform for analyzing oligonucleotide delivery, allowing customization of chemical structures for specific purposes or targets. As such, this platform will maximize the potential of oligonucleotide therapeutics.
Acknowledgment
We would like to express our sincere gratitude to Professor Akimitsu Okamoto, Associate Professor Kunihiko Morihiro and Dr. Masako Takatsu of the Graduate School of Engineering, the University of Tokyo for their great cooperation and generous advice.
Dr. Akimitsu Okamoto lab website: https://webpark1516.sakura.ne.jp/
Reference
M. Takatsu, K. Morihiro, H. Watanabe, M. Yuki, T. Hattori, K. Noi, K. Aikawa, K. Noguchi, M. Yohda, T. Okazoe, and A. Okamoto, Cellular penetration and intracellular dynamics of perfluorocarbon-conjugated DNA/RNA as a potential means of conditional nucleic acid delivery, ACS Chem. Biol. 18(12), Nov 19, 2023, 2590-2598.
Product information
AX/AX R with NSPARC Confocal-based Super Resolution Microscope
The super-resolution detector NSPARC features an array of 25 subunit detectors to achieve higher resolution and signal-to-noise ratio without comprising the functionality of the conventional AX/AX R confocal microscope.
- Home
- intergrundwissen
- Application Notes
