- zh Change Region
- Global Site
- 主页
- CRO
- Shonan Health Innovation Park
- Applications & Technology


Nikon BioImaging Labratories
Shonan Health Innovation Park
Applications & Technology
Provided here is an overview of the various applications supported by the Nikon BioImaging Lab, detailing imaging, analysis, and other tools we have at our disposal. Click to jump to a specific application, or scroll down to start exploring.
The information on these pages applies to the Nikon BioImaging Lab in Shonan, Japan. There are also laboratories in Leiden, The Netherlands and Cambridge, Massachusetts, USA. For more information, please contact the facility nearest to your location.
High Content Imaging and Analysis
Technology: Confocal Microscope Systems, ECLIPSE Ti2-E Inverted Microscope, NIS-Elements software, NIS.ai deep learning software
High content imaging is about imaging more – more detection channels, more time points, more data. It’s no wonder that high content imaging and analysis is a powerful tool for so many applications, such as phenotypic screening in drug discovery research. However, the greatest strength of high content imaging – the volume of rich data created – can also be its greatest drawback, creating critical bottlenecks in experiment design, data storage, and image analysis. At the Nikon BioImaging Lab, our high content imaging services are centered around a stable yet flexible platform for long-term live-cell high content that address common process bottlenecks to provide operational efficiency.
High content imaging with the BioPipeline LIVE is great for applications ranging from cell health monitoring, to toxicology studies, and more and more. For imaging thicker samples, a Nikon resonant scanning confocal system acquiring data deep in samples such as spheroids, organoids, organ-on-a-chip, and tissue. This system also features our NIS.ai deep learning software modules, which can be trained to perform a number of different analysis tasks, such as segmentation, and can be integrated into an automated imaging and analysis pipeline.
3D Imaging
Technology: Confocal Microscope Systems, ECLIPSE Ti2-E Inverted Microscope
Cell biology and other fields have long relied on in vitro imaging of a single layer of cultured cells adhering to a flat surface, but the limitations of such model systems are being increasingly recognized, and more physiologically-relevant models are gaining in popularity, including spheroids, organoids, organ-on-a-chip, and similar systems. While these systems provide better models of organismal biology, they have non-trivial thickness and 3D structure, which limits the application of routine 2D widefield imaging approaches.
Our flagship point-scanning confocal system is capable of deep imaging tens of micrometers or more into the sample. Our Ti2-E inverted microscope is also equipped for 3D widefield imaging, featuring integrated deconvolution software for achieving optical sectioning in samples to approximately 20 micrometers thick. While 3D imaging by widefield deconvolution doesn’t provide the instantaneous 3D sectioning of a confocal instruments, it is fast and often the superior choice for low-signal samples. We understand the nuances that inform proper choice of 3D imaging technique, and are here to make sure you are successful.
Z-stack of a kidney organoid acquired using a Nikon A1R HD25 confocal system.
Red: Actin cytoskeleton (filamentous-actin)
Blue: Nuclei (DNA)
Fluorescence Imaging
Technology: Confocal Microscope Systems, ECLIPSE Ti2-E Inverted Microscope
Fluorescence microscopy is widely considered to be one of the most sensitive imaging methods for detecting a given molecular target with high specificity. Fixed samples are generally labeled with dye-conjugated antibodies using standard immunohistochemistry/immunofluorescence techniques. Live samples can be made to genetically express a fluorescent protein fused to a target protein of interest or stained with membrane-permeable organic dyes.
The A1R HD25 features laser illumination, and the Ti2-E inverted microscope feature LED illumination. Specifically, our Ti2-E features a fast color-switching Lumencor Spectra-X 7-channel LED light engine for advanced multiplexing experiments. The A1R HD25 is configured with 6 different color laser lines, including 445 nm and 514 nm lines, which are commonly used for CFP/YFP FRET studies. It is also capable of performing targeted FRAP and photostimulation experiments. We stand ready with the expertise to craft successful fluorescence-based assays, having a deep knowledge of fluorophores, microscope settings, mounting media, and related concerns.
Mouse hippocampal neuron. Green: GFP (dendrites), Red: PSD-95-TagRFP (postsynaptic marker). Image acquired using a Nikon CFI SR HP Apochromat TIRF 100XC Oil objective and courtesy of: Drs. Yutaro Kashiwagi and Shigeo Okabe, Department of Cellular Neurobiology, Graduate School of Medicine and Faculty of Medicine, The University of Tokyo.
High-Resolution Imaging
Technology: A1R HD25 Confocal, Ti2-E Inverted Microscope
It’s easy to find a microscope providing high magnification, but it takes much more than magnification alone to acquire quality high-resolution images. Our A1R HD25 confocal system provides both optical sectioning – and high-resolution imaging of sample features in that 2D section. To maximize the resolution of our confocal instrument, we have the A1-ER HD25 data, which includes detailed point-spread function (PSF) models for Nikon’s market-leading objective lenses.
The Ti2-E inverted microscope is also capable of high-resolution multi-dimensional imaging – able to acquire data in 3D, over time, and in multiple color channels similar to our confocal system – and includes deconvolution software for maximizing resolution and optical sectioning.
It’s not just the microscope that goes into making high resolution images, equally important is proper sample preparation. If, for example, your sample is mounted in a medium with the wrong refractive index, optical resolution will suffer significantly, especially in the Z direction. At the Nikon BioImaging Lab we have you covered on both fronts, with expertise in both high-resolution microscopy and sample preparation.
Cochlear cilia XY and XZ views before (left) and after (right) deconvolution using the A1-ER enhanced resolution module. Image data acquired using a Nikon A1R with a pinhole diameter of 1 Airy Unit.
Color Imaging
Technology: ECLIPSE Ti2-E Inverted Microscope
Our Ti2-E inverted microscope is equipped with a Lumencor LIDA light source for transmitted light (diascopic) illumination. The LIDA consists of individual red, green, and blue LEDs that can be triggered in succession, imaging each color sequentially, and combining the three images into a composite RGB color image. This is an advantage as it allows us to avoid using single exposure color cameras, which have lower sensitivity, spatial resolution, and dynamic range than their monochrome counterparts. Furthermore, the intensity of each LED color is individually addressable, unlocking very fine hardware-based control of color balancing without image post-processing or having to move any system components.
This technology is beneficial not only for pathologists and histologists, but to anyone looking for a modern approach to quantitative color imaging. This is even beneficial for live cell imaging applications as the LIDA does not feature any UV or near-IR light in its spectrum. In addition to brightfield, other techniques we support that can benefit from color imaging include polarized light, differential interference contrast (DIC), and darkfield microscopies. As always, the Nikon BioImaging Lab staff is here to advise you on best practices for your entire microscopy imaging and analysis workflow, including color imaging.
Brightfield color image of an H&E stained mammalian skeletal muscle tissue section acquired using a Nikon CFI Plan Apo Lambda 10X objective.
Label-free Imaging
Technology: BioStudio-T, ECLIPSE Ti2-E Inverted Microscope, NIS.ai
While fluorescence imaging might be one of the most popular contrasting technique for optical microscopy, it is also one of the most invasive. Fluorophores are prone to photobleaching, limiting their useful lifetime, and resulting in the need for greater and greater illumination intensity to excite a similar amount of emission as the experiment progresses. This difficulty is compounded by the phototoxic response of live cells to high intensity illumination. Assays using sensitive cell types, such as stem cells, may not be completely compatible with fluorescence imaging of the desired target.
Cell health concerns are helping to fuel the renewed popularity of label-free imaging techniques, which help avoid both the need for high intensity illumination as well as perturbation of the system by the labeling process.
Artificial intelligence-based machine learning and deep learning analysis techniques are also helping to enable this trend. For example, the Convert.ai module (one of several NIS.ai deep learning software modules) automates the identification of key cell features from differential interference contrast (DIC) data, such as the simulated DAPI staining in the figure here. All of our systems support label-free imaging, with available techniques including new volume contrast, phase contrast, differential interference contrast (DIC), darkfield, and brightfield. Contact us today to see if label-free imaging will work for you.

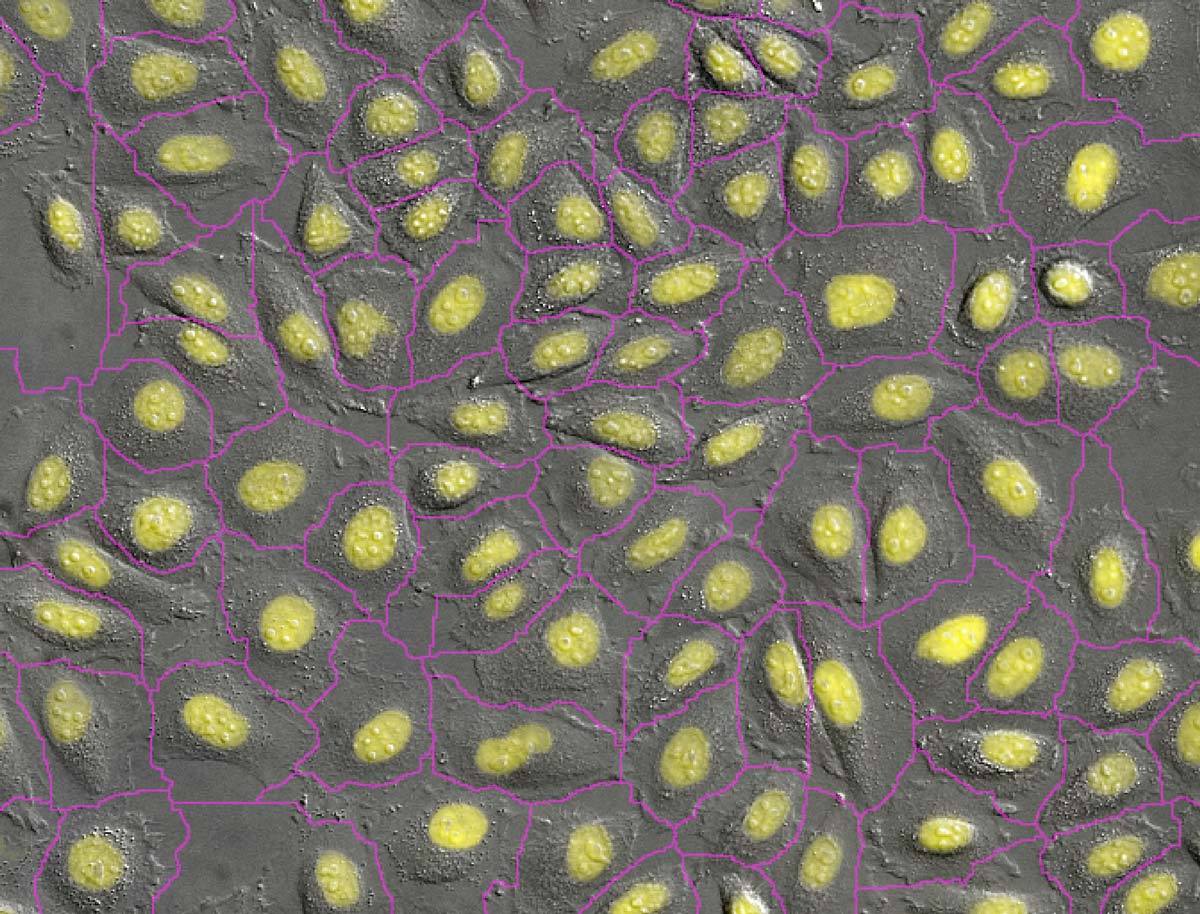
DAPI标记细胞核是细胞计数与识别的常用方法。仅需DIC或相差图像Convert.ai即可用于训练推测DAPI图像。生成的推测图像即可用于图像分割与计数,您无需标记DAPI也不需要拍摄荧光图像。
照片来源:北海道大学电子科学研究所技术学院的Kentaro Kobayashi博士
Cell Screening
Technology: BioStudio-T
Stem cells are essential tools in regenerative medicine and allied fields, however they can be notoriously difficult to culture and maintain in the undifferentiated state due to their high environmental sensitivity. To address the challenges of culturing stem cells (and other difficult cell types), we operate a Nikon BioStudio-T.
The BioStudio-T is a compact single-vessel microscope imaging system that fits within an existing incubator. Unlike other microscopes, the sample is held stationary while the objective lens is scanned, reducing disturbance to the sample.
相差图像的无标记分析
红色=未分化
绿色=已分化
- 主页
- CRO
- Shonan Health Innovation Park
- Applications & Technology
