- en Change Region
- Global Site
Application Notes
Protein components of virus particles in SARS-CoV-2-infected cells imaged by N-STORM super-resolution microscope
November 2023
Elucidating the mechanism of SARS-CoV-2 proliferation is important for the development of prevention and treatment strategies for COVID-19. In cells infected with SARS-CoV-2, the expression levels, ratios, and intracellular localizations of each virus-constituting protein change as virus particles are formed. Although endosomes with numerous intraluminal vesicles (multivesicular bodies; MVBs) and lysosomes are now being recognized as locations where virus particles are assembled, it is unclear how each viral protein accumulates within them.
Ms. Yoko Ishida, Dr. Shinichiro Okamoto, Dr. Megumu Takahashi, and Prof. Hiroyuki Hioki (Department of Neuroanatomy, Graduate School of Medicine, Juntendo University) observed the localization of the spike (S) protein and nucleocapsid (N) protein within MVBs and lysosomes using both a confocal microscope and N-STORM super-resolution microscope.
In this application note, we present images of virus particle protein components in SARS-CoV-2-infected cells successfully captured by N-STORM.
Keywords: SARS-CoV-2, COVID-19, spike (S) protein, nucleocapsid (N) protein, N-STORM super-resolution microscope
Overview
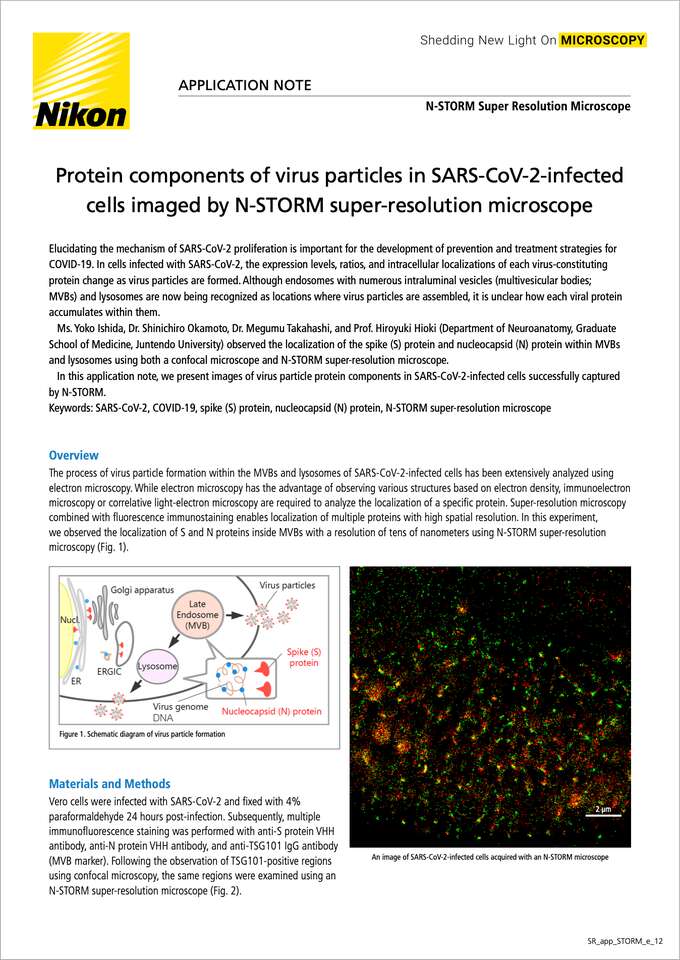
The process of virus particle formation within the MVBs and lysosomes of SARS-CoV-2-infected cells has been extensively analyzed using electron microscopy. While electron microscopy has the advantage of observing various structures based on electron density, immunoelectron microscopy or correlative light-electron microscopy are required to analyze the localization of a specific protein. Super-resolution microscopy combined with fluorescence immunostaining enables localization of multiple proteins with high spatial resolution. In this experiment, we observed the localization of S and N proteins inside MVBs with a resolution of tens of nanometers using N-STORM super-resolution microscopy (Fig. 1).
Figure 1. Schematic diagram of virus particle formation
An image of SARS-CoV-2-infected cells acquired with an N-STORM microscope
Materials and Methods
Vero cells were infected with SARS-CoV-2 and fixed with 4% paraformaldehyde 24 hours post-infection. Subsequently, multiple immunofluorescence staining was performed with anti-S protein VHH antibody, anti-N protein VHH antibody, and anti-TSG101 IgG antibody (MVB marker). Following the observation of TSG101-positive regions using confocal microscopy, the same regions were examined using an N-STORM super-resolution microscope (Fig. 2).
N-STORM
Figure 2. Images of SARS-CoV-2-infected cells acquired with a confocal microscope and an N-STORM super-resolution microscope
(A) Image captured with a confocal microscope
Confocal system: A1R HD25, Objective: CFI Apochromat Lambda S 60X Oil, NA = 1.4
(B) Image of the same field of view as panel A captured with N-STORM super-resolution microscope
(C) Enlarged image of the region highlighted in panel B
Objective: CFI SR HP Apochromat TIRF 100XC Oil, NA = 1.49
Green: S protein (ATTO647), Red: N protein (Alexa Fluor 568)
Results
Observation of the infected cells with a confocal microscope revealed that most of the fluorescence signals for S and N proteins were colocalized (Fig. 2A). Observation of the same field using an N-STORM super-resolution microscope elucidated the intracellular localization of S and N proteins at higher spatial resolution (Fig. 2B). In the enlarged image, the fluorescence signals generated by the immunolabeled S and N proteins appear as separate but proximally-located bright spots (Fig. 2C).
Summary
Super-resolution microscopy is a technique that enables observation of detailed structures beyond the diffraction limit of light, and it achieves optical observation of ultra-fine morphologies that could previously only be observed with electron microscopy. Furthermore, N-STORM makes it possible to perform super-resolution localization of multiple antigens within the same sample. In this study, we observed the intracellular distribution of S and N proteins within SARS-CoV-2-infected cells at high spatial resolution. We have demonstrated the capability of N-STORM toward analyzing the intracellular localization of virus particle protein components with high precision, thus contributing to the elucidation of the virus particle formation mechanism.
Acknowledgments
We would like to express our sincere gratitude to Ms. Yoko Ishida, Dr. Shinichiro Okamoto, Dr. Megumu Takahashi, and Prof. Hiroyuki Hioki(Department of Neuroanatomy, Graduate School of Medicine, Juntendo University) for their generous cooperation in providing images. We would also like to appreciate Prof. Atsushi Kawaguchi (Department of Infection Biology, Faculty of Medicine, University of Tsukuba) for providing the Vero cell specimens infected with SARS-CoV-2.
This is part of the research results donated by Nikon Corporation for efforts to conduct advanced image analysis using optical microscopes as an aid to basic research on viral infectious diseases.
References
Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress.
Mendonça et al.
Nat. Commun., 2021;12(1):4629.
https://www.nature.com/articles/s41467-021-24887-y
SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress.
Scherer et al.
Sci. Adv., 2022;8(1):eabl4895.
https://www.science.org/doi/10.1126/sciadv.abl4895
Product information
N-STORM Super Resolution Microscope
The N-STORM utilizes a localization technique called STochastic Optical Reconstruction Microscopy (STORM) to achieve a resolution of ten times that of conventional light microscopes. It allows imaging of the structure of cell organelles on a molecular level.
- Lateral resolution: Approx. 20 nm
- Axial resolution: Approx. 50 nm
