Romain Le Bars, Ph.D.
The Imagerie-Gif light microscopy core facility is a member of the France Bioimaging Infrastructure. The facility is hosted by the Institute for Integrative Biology of the Cell (I2BC) at Gif sur Yvette, France.
Equipamento Nikon
- N-STORM Super-Resolution
- Ti-E Inverted Microscope (see current model)
Please introduce your imaging facility and summarize your research.
The Imagerie-Gif light microscopy core facility is a member of the France Bioimaging Infrastructure. The facility is hosted by the Institute for Integrative Biology of the Cell (I2BC) at Gif sur Yvette, France. Our users are mainly interested in the observation of the sub-cellular organisation and processes, over a broad diversity of models organisms (bacteria, yeast, animal cells, plants…).
We started to use super resolution techniques such as PALM and STORM in 2014 to study the mechanisms involved in the organisation of the bacterial chromosome of E. coli and V. cholerae. At this resolution we were able to precisely localise the distribution of multiple proteins involved in the choreography of the chromosome organisation along cell division. Based on the single molecule localisation microscopy approach, we developed methods to quantify the level of protein clustering and performed single particle tracking (SPT-PALM) to measure protein-DNA interaction. Since we adapted the protocols to investigate the endoplasmic reticulum network in plants and cytoskeleton organisation in animal cells.
Why use N-STORM systems?
We chose the Nikon N-STORM system to investigate our subjects for the robustness and the quality of their instruments. This system assures a high stability and a very good precision for those nanoscale observations by the use of the Perfect Focus System to prevent any drift of the sample and also uses two approaches to correct drifting: either by autocorrelation or by imaging fiducial markers.
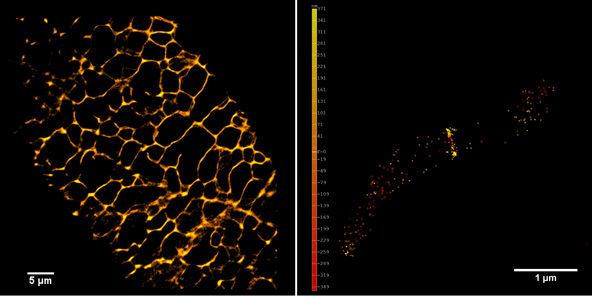
Left panel: The plant endoplasmic reticulum network observed in PALM on living Arabidopsis thaliana root cell expressing Calnexin-mEOS.
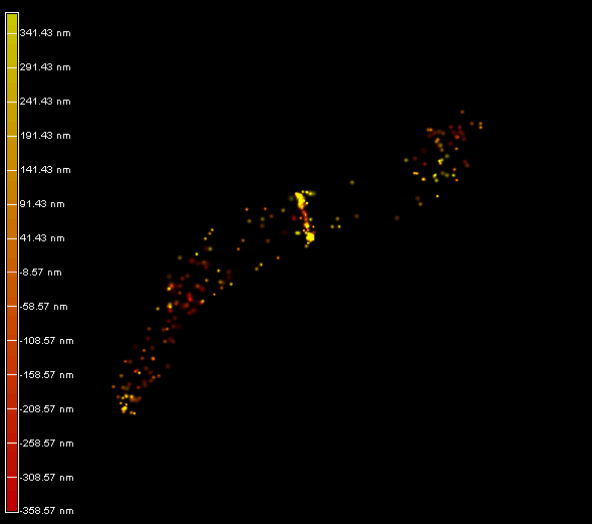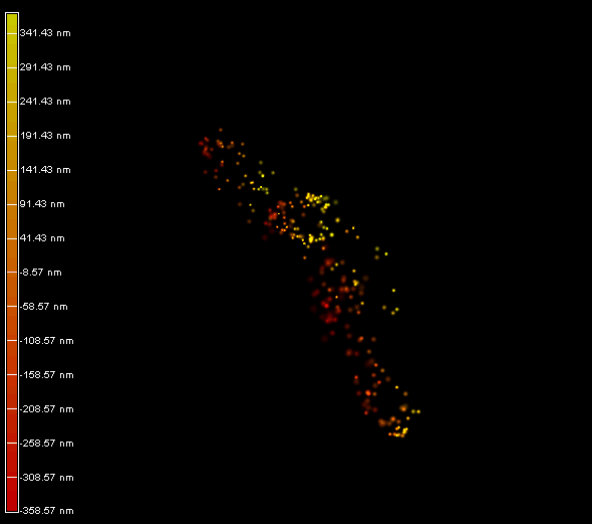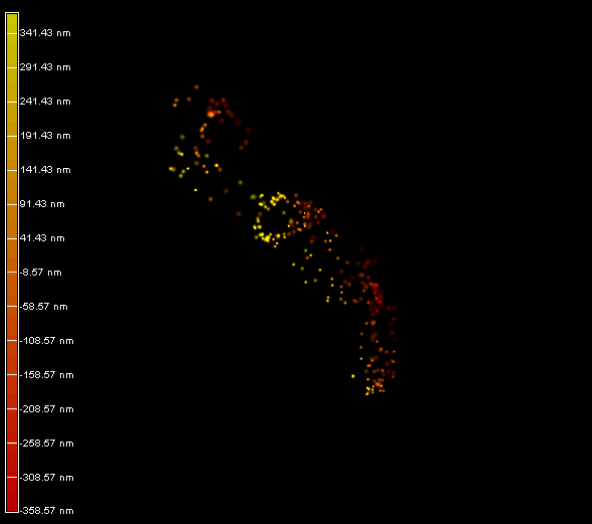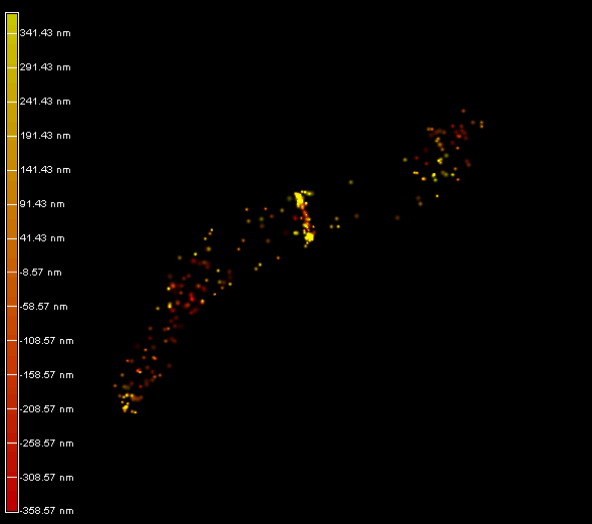
Right panel and movie: 3D-PALM imaging of the FtsZ protein coupled with mEOS2 in Vibrio cholera.
Ref: Galli, E. et al. Cell division licensing in the multi-chromosomal Vibrio cholerae bacterium. Nat Microbiol 1, 16094 (2016).
