- pt Change Region
- Global Site
Notas de aplicação
High-resolution macro-to-micro 3D observation of cleared rice anthers : sample preparation in view of the refractive index of immersion media
abril 2023
Flowers have organs called anthers. This organ, also commonly called the “stamen”, has a bag-like structure in which pollen develops. Pollen formation is a process of a succession of generations accompanied by meiosis, and the generation and development of anthers as well as pollen have important biological implications. Anther generation and development have long been observed using optical and electron microscopes, and in recent years research has been conducted at the genetic and molecular level. Most recently, microRNA-mediated regulation of anther wall (anther sack) formation has been revealed and attracted attention (Komiya et al.).
One microscope observation technique that has been attracting attention in recent years is a technique for making transparent tissues, organs, and individuals. Techniques such as ClearSee, TOMEI, and PEA-CLARITY have been developed to make transparent plants, and ClearSeeAlpha (Kurihara et al.) and iTOMEI (Sakamoto et al.) are further developments thereof. These clearing reagents are often used in combination with confocal microscopy and multiphoton excitation microscopy, which are effective for the deep observation of samples.
Conventionally, anther development was observed in sections prepared using a microtome, but it is difficult to understand accurate three-dimensional structures from slices of two-dimensional images. In this respect, the combination of clearing method and confocal microscopy is considered to be very effective for 3D observation of deep sites. However, there have been problems concerning the degree of clearing and processing time of the clearing reagent differing according to the plant species and types of tissues/organs, as well as the inevitability of aberrations accompanying deep observation with a microscope.
This application note introduces an example of acquisition of 3D images of rice anthers under optimal optical conditions in order to understand their structures at the cellular level, by searching for the best clearing reagent, applying clearing, and mounting the samples with a solidified clearing reagent.
Rice ears were collected from rice (Nipponbare) grown under long daylight conditions and fixed with 4% (w/v) paraformaldehyde. After removing the anthers by stripping the lemma and palea under a stereomicroscope, the anthers were stained with 0.01% (v/v) SR2200 (mainly for staining cell walls) and 0.1 µg/ml Propidium Iodide (for staining nuclei as well as cell walls). Then, they were immersed in ClearSee, ClearSeeAlpha (50 mM sodium sulfite), iTOMEI, RapiClear1.47, RapiClear1.52, and 45% (v/v) RapiClear1.52 for 2-3 days to render them transparent.
Among the clearing reagents used in this experiment, ClearSee, ClearSeeAlpha, and iTOMEI resulted in well-shaped anthers without contraction of the anther locule (Table 1). The cleared anthers were first mounted in a 1% agarose gel made with 53% (v/v) glycerin water (refractive index: 1.41), but they seemed to revert from clear back to their original color. Therefore, by adding various concentrations of agarose with the aim of solidifying ClearSeeAlpha and iTOMEI, it was found that soft gels can be made with 2% (w/v) in ClearSeeAlpha and with 1%(w/v) in iTOMEI.
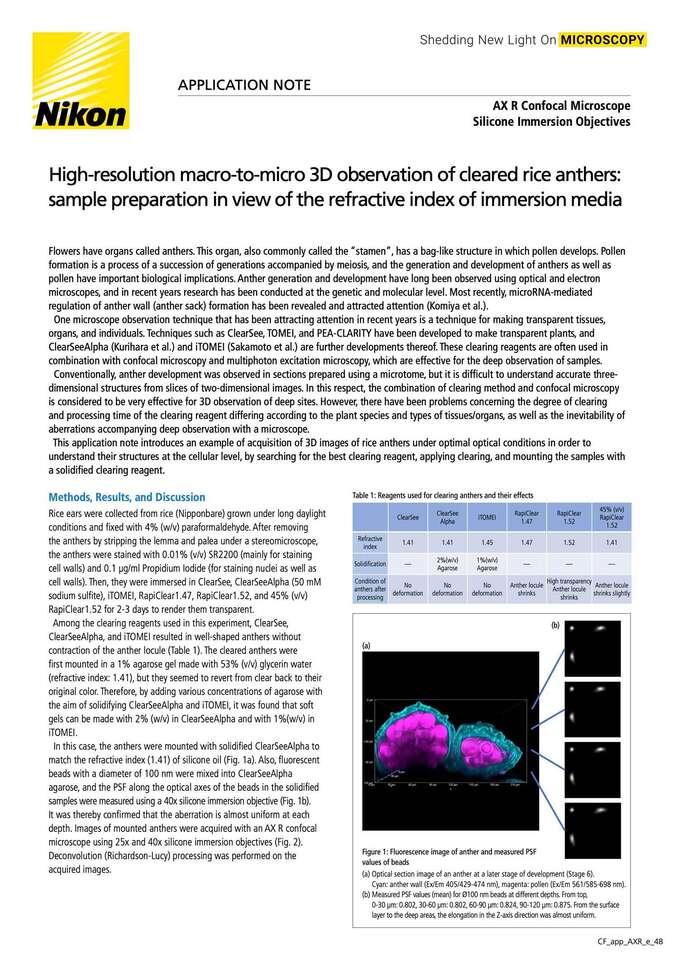
Figure 1: Fluorescence image of anther and measured PSF values of beads
(a)Optical section image of an anther at a later stage of development (Stage 6).
Cyan: anther wall (Ex/Em 405/429-474 nm), magenta: pollen (Ex/Em 561/585-698 nm).
(b)Measured PSF values (mean) for Ø100 nm beads at different depths. From top,
0-30 µm: 0.802, 30-60 µm: 0.802, 60-90 µm: 0.824, 90-120 µm: 0.875. From the surface layer to the deep areas, the elongation in the Z-axis direction was almost uniform.
Methods, Results, and Discussion
In this case, the anthers were mounted with solidified ClearSeeAlpha to match the refractive index (1.41) of silicone oil (Fig. 1a). Also, fluorescent beads with a diameter of 100 nm were mixed into ClearSeeAlpha agarose, and the PSF along the optical axes of the beads in the solidified samples were measured using a 40x silicone immersion objective (Fig. 1b). It was thereby confirmed that the aberration is almost uniform at each depth. Images of mounted anthers were acquired with an AX R confocal microscope using 25X and 40X silicone immersion objectives (Fig. 2). Deconvolution (Richardson-Lucy) processing was performed on the acquired images.
Whole image of cleared anther. Cyan: anther wall, magenta: pollen.
Cleared with ClearSeeAlpha, mounted in gels made with this clearing solution, and imaged with 1 x 4 tiling and Z-stack. With a 25x silicone immersion lens and large field of view, high- resolution images of an entire anther that has developed to about 1 x 0.3 x 0.25 mm were obtained.
Objective: CFI Plan Apo λS 25XC Sil, Zoom: 1x, Pixel size: 0.35 µm
Enlargement of the white frame in Fig. a.
With a higher NA objective, the contours of epidermal cells can be clearly observed. Optical cross-section images show that the shape of epidermal cells and pollen can be observed to deep areas with minimal deformation.
Objective: CFI Plan Apo λS 40XC Sil, Zoom: 2x, Pixel size: 0.21 µm
Enlargement of the white frame in Fig. b. Imaging at high magnification using a 40x silicon immersion objective allows acquisition of surface structures and 3D images of epidermal and endothelial cells (arrows).
Zoom: 9x, Pixel size: 0.05 μm
Figure 2: Rice anther captured from macro to micro with a silicone immersion objective
Summary/Outlook
Rice anthers cleared with ClearSeeAlpha with a refractive index of 1.41 are mounted in a gel made with this clearing reagent, and a silicon immersion objective is utilized to enable observation almost free from aberrations caused by differences in refractive index. In the Z direction, highly-precise observation at the cellular level is possible at depths of up to about 200 µm.
Such observation eliminates the need to prepare sections with paraffin or resin, and facilitates spatiotemporal analysis of development and cell differentiation.
In the future, it is expected that quantification of cell shapes and volumes will be realized by observing the intracellular structures of deep cells using a silicone immersion objective with higher NA and resolution, and analyzing images acquired with AI-equipped software.
Acknowledgements
We would like to express our sincere gratitude to Dr. Reina Komiya of the Science Technology Group, Okinawa Institute of Science and Technology Graduate University, and Dr. Koji Koizumi of the Imaging Section of the Okinawa Institute of Science and Technology Graduate University for providing specimens and images.
References
Araki S, Le NT, Koizumi K, Villar-Briones A, Nonomura KI, Endo M, Inoue H, Saze H, Komiya R (2020). miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat Commun. 11: 3115.
Kurihara D, Mizuta Y, Nagahara S, Higashiyama T (2021). ClearSeeAlpha: Advanced Optical Clearing for Whole-Plant Imaging. Plant Cell Physiol. 62: 1302-1310.
Sakamoto Y, Ishimoto A, Sakai Y, Sato M, Nishihama R, Abe K, Sano Y, Furuichi T, Tsuji H, Kohchi T, Matsunaga S (2022). Improved clearing method contributes to deep imaging of plant organs. Commun Biol . 5: 12.
Product Information
The 3D structure of cleared specimens , which have a refractive index close to that of silicone oil, can be captured brightly and with high definition, down to their deepest areas. When used with a large field-of-view AX confocal microscope and a high-speed resonant scanner, they can efficiently acquire images of an entire specimen.
