Dr. Steve Thomas
Senior Lecturer in Cardiovascular Science, University of Birmingham
尼康设备
Please tell us about your research.
We are interested in platelets and megakaryocytes, which are blood cells. Particularly, we study how they are formed, how they function both in normal healthy people but also in disease. We use a lot of imaging to try to learn what is happening at a cellular level, understand the proteins that are involved in making the cells work, how they interact together and what happens when these things go wrong in terms of human disease.
What imaging techniques are you using and how do these imaging techniques help you answer your research questions?
We are using fluorescence microscopy, predominantly super resolution techniques. These are really important for us because they allow us to see the sub-cellular detail that is normally excluded when you are using diffraction limited techniques. Super-resolution has allowed us to really look at interactions between different protein compartments in the cell. We can see how they are organized spatially which tells us something about how they function and what they are doing in the cell. We can’t do that in normal microscopy which does not have the resolution. Both the N-STORM and the N-SIM techniques have been very useful in understanding how these proteins are organized and how that dictates their function in the cell.

You mentioned that you use N-STORM and N-SIM – what did each of these two techniques enable you to see particularly?
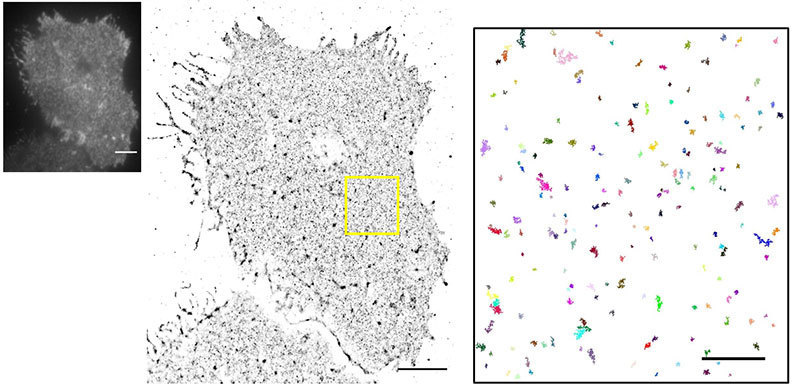
N-STORM allows us to look at individual molecules within the platelets surface and to try to understand how they are organized. For example, we may label a receptor on that platelet surface and look at how its distribution changes in an activated vs. a resting cell. That can tell us something about how these molecules are helping this cell to signal because we can get information on cluster size and number of molecules, see how they co-localize with other molecules in the cell. Insight into signaling on a molecular level feeds into drug discovery and understanding of the disease by knowing what the normal situation is vs what happens if these processes go wrong. So N-STORM specifically allows us to check what is happening with these proteins at the molecular level.
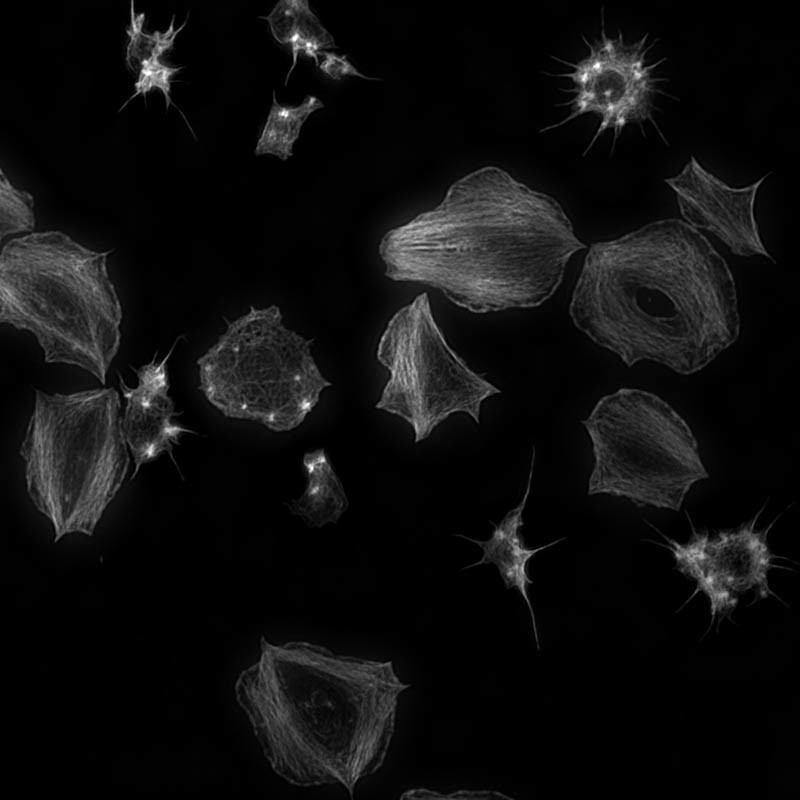
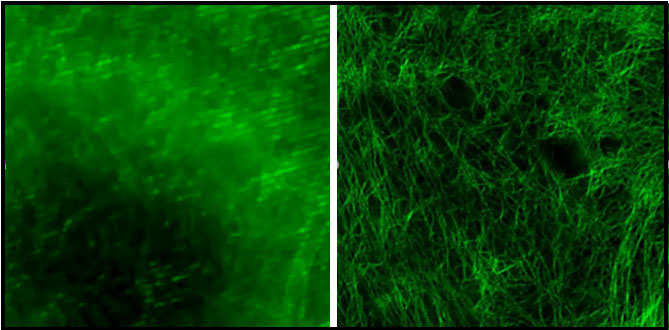
N-SIM on the other hand is very nice for giving us high-quality images of the cytoskeleton, which is a protein network in the cell. This allows us to look at how this protein network is organized in a normal cell and what happens when you activate these proteins and to drive some of the cell functions. For example, when the platelets are activated they change shape very rapidly. By imaging the cytoskeleton using N-SIM we can understand how these proteins are responsible for that changing shape.
You mentioned that you use N-STORM and N-SIM – what did each of these two techniques enable you to see particularly?
N-STORM allows us to look at individual molecules within the platelets surface and to try to understand how they are organized. For example, we may label a receptor on that platelet surface and look at how its distribution changes in an activated vs. a resting cell. That can tell us something about how these molecules are helping this cell to signal because we can get information on cluster size and number of molecules, see how they co-localize with other molecules in the cell. Insight into signaling on a molecular level feeds into drug discovery and understanding of the disease by knowing what the normal situation is vs what happens if these processes go wrong. So N-STORM specifically allows us to check what is happening with these proteins at the molecular level.
N-SIM on the other hand is very nice for giving us high-quality images of the cytoskeleton, which is a protein network in the cell. This allows us to look at how this protein network is organized in a normal cell and what happens when you activate these proteins and to drive some of the cell functions. For example, when the platelets are activated they change shape very rapidly. By imaging the cytoskeleton using N-SIM we can understand how these proteins are responsible for that changing shape.
What do you think of Nikon microscopes?
The quality of the systems is very good, they are well-built and well-made, but primarily I think the support that we get for using the systems is excellent and really helps us to make the most out of our systems. You have got people who really understand them and know how to use them to solve questions. The support behind the systems is very important.
What do you think of Nikon microscopes?
The quality of the systems is very good, they are well-built and well-made, but primarily I think the support that we get for using the systems is excellent and really helps us to make the most out of our systems. You have got people who really understand them and know how to use them to solve questions. The support behind the systems is very important.
What are the next steps in your research and how will your N-SIM-S system support it?
I think the increase in speed of the N-SIM-S over the older systems and the fact that we have got the two camera set up so we can do multicolour simultaneous [imaging] means we can now start to actually look in live cells at two different components and see how they interact together. I think that is going to really open up a lot of the work for us because we can start to not just look at one thing or another in fixed cells. Instead, we can start to do this over time and see how the different components interact, how they influence each other. The ability to do this rapidly over time in two colors is really going to allow us to look at more complex systems.
What are the next steps in your research and how will your N-SIM-S system support it?
I think the increase in speed of the N-SIM-S over the older systems and the fact that we have got the two camera set up so we can do multicolour simultaneous [imaging] means we can now start to actually look in live cells at two different components and see how they interact together. I think that is going to really open up a lot of the work for us because we can start to not just look at one thing or another in fixed cells. Instead, we can start to do this over time and see how the different components interact, how they influence each other. The ability to do this rapidly over time in two colors is really going to allow us to look at more complex systems.
